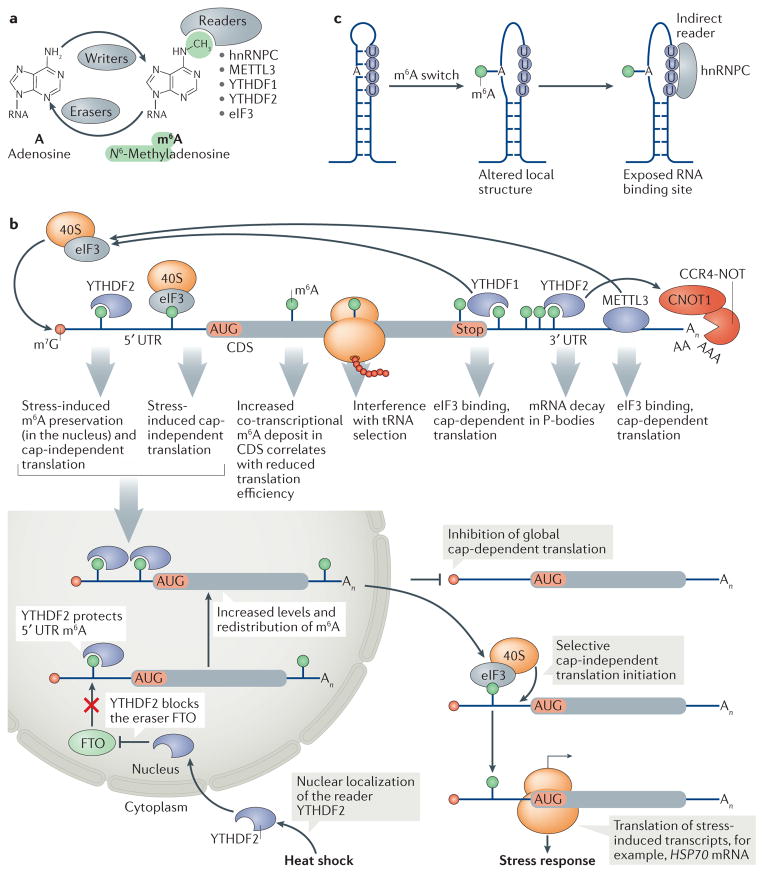
Figure 4. The effects of N6-methyladenosine on mRNA translation and decay.
a | ‘Writer’ proteins establish N6-methyladenosine (m6A) at internal RNA sites, ‘eraser’ proteins remove them, and ‘reader’ proteins directly bind the N6-methyl group of m6A (reviewed in REF. 148). The listed readers affect the translation and stability of m6A-modified mRNAs. b | m6A modifications in mRNAs occur with a preference for the last exon and 3′ untranslated region (UTR)201 and are increased during stress152. According to its position in the mRNA, m6A is bound by readers that can induce cap-dependent translation or internal ribosome recruitment. In the 5′ UTR, eukaryotic initiation factor 3 (eIF3) can directly bind m6A and facilitate internal translation initiation151. Stress-responsive 5′ UTR N6-adenosine methylation, for example during heat shock, is preserved by YTH domain-containing family protein 2 (YTHDF2), which blocks binding of the eraser fat mass and obesity-associated protein (FTO), thereby promoting cap-independent translation initiation of stress response mRNAs152. m6A in the coding sequence (CDS) is linked to tRNA selection163, and at the 3′ UTR it is linked to increased translation owing to YTHDF1 binding to m6A and eIF3 recruitment for cap-dependent translation158. The writer methyltransferase like 3 protein (METTL3) can also directly bind to eIF3 to increase translation of m6A-containing mRNAs independently of its m6A writer activity159. By contrast, YTHDF2 promotes degradation of m6A-modified mRNAs by recruiting the deadenylase complex CCR4–NOT157. Together, increased translation efficiency and activated decay of m6A-modified mRNAs allow dynamic regulation of protein synthesis. c | m6A mRNA modifications are associated with unfolded RNA structures. N6 adenosine methylation in a stem disrupts based paired regions (‘m6A switch’), which allows binding of the ‘indirect reader’ heterogeneous nuclear ribonucleoprotein C (hnRNPC) to exposed U rich motifs in the nucleus153. CNOT1, CCR4–NOT transcription complex subunit 1; HSP70, heat shock protein 70; P body, processing body. Part c is adapted from REF. 153, Macmillan Publishers Limited.