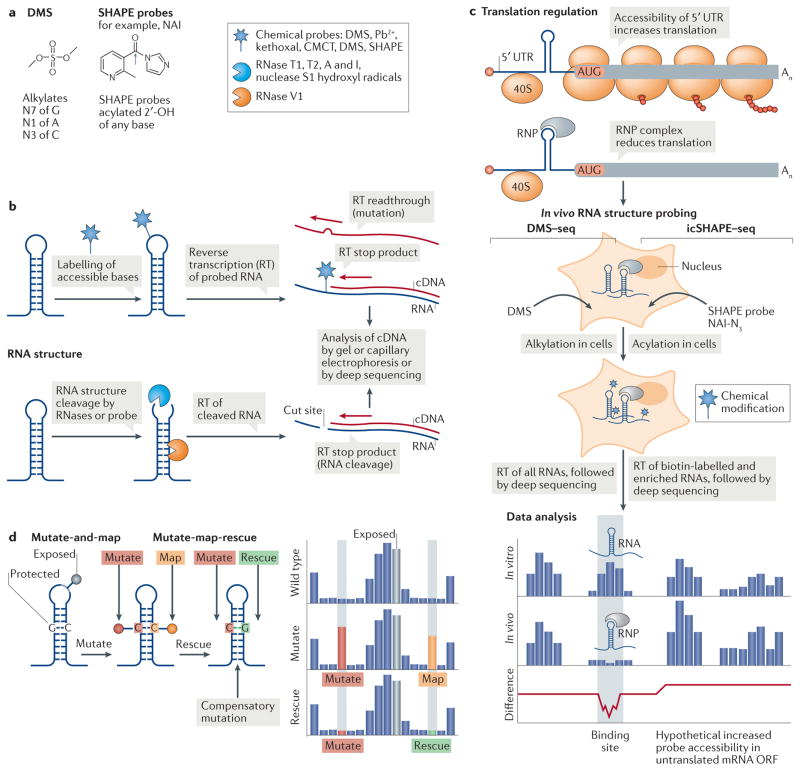
Figure 5. Global RNA structure probing to assess translation regulation.
Global RNA structure probing inside cells can assess the transcriptome structure in the presence of proteins. a | Chemical schematics of the RNA structure probes dimethyl sulfate (DMS) and the selective 2′-hydroxyl acylation analysed by primer extension (SHAPE) reagent 2 methylnicotinic acid imidazolide (NAI) and their reactivity. The grey arrow indicates the site of 2′-OH attack of the RNA by the probe. Different probes induce single-strand-specific chemical labelling (star) or cleavage by enzymes or probes (blue Pac Man shape, single strand specific; orange Pac Man, double strand specific). b | RNA probing either labels RNA through a covalent reaction of a chemical probe with accessible nucleotides (top) or cleaves the RNA backbone with RNases (bottom). A pool of modified or cleaved RNAs is transcribed into cDNA by reverse transcription (RT), and modified or cleaved sites are identified by their effect on RT. c | A 5′ untranslated region (UTR) ribonucleoprotein (RNP) complex that inhibits ribosome scanning reduces the accessibility of the mRNA for the probe in cells. In in vivo RNA structure probing, shown here for DMS treatment followed by deep sequencing (DMS–seq) and in vivo click SHAPE followed by deep sequencing (icSHAPE–seq), RNA structures areprobedincells bychemicalmodification. Data analysis of in vitro-probed and in vivo-probed RNA can indicate the presence of RNA structures as protein binding sites, owing to masked probe accessibility or to remodelling of the structure by protein interaction. Where a 5′ UTR RNP complex inhibits the translation of an open reading frame (ORF), the accessibility of the probe to poorly translated coding sequences might increase in the absence of ribosomes. d | In multidimensional mutate and map chemical probing, a mutation (red) that eliminates base pairing exposes the mutated nucleotide and its partner nucleotide (orange) to chemical modification (pins). In mutate-map-rescue probing, a mutation is rescued from modification by a compensatory mutation of the partner nucleotide (green). The reactivity profile reflects changes in probe accessibility upon mutation (red), which allows mapping (orange) of the base-paired nucleotide, while rescue (green) confirms base pairing. Nucleotides in loops are exposed and accessible to the probe. Pb2+, lead; CMCT, 1 cyclohexyl (2-morpholinoethyl)carbodiimide metho-p-toluene sulfonate.