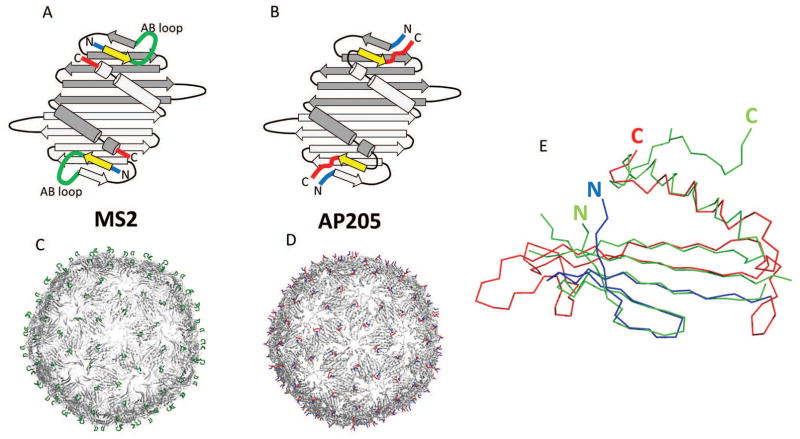
Figure 2. Structural features of bacteriophage AP205 coat protein.
Coat protein in AP205 and related phages, such as MS2, builds very stable dimers. Two monomers are shown in different shades of grey (panels A and B). Notice the close proximity of N- (blue) and C- (red) termini in dimers. 90 dimers further assemble into VLPs (panels C and D). In MS2, AB loop (green) is the most exposed structure on the surface of VLPs. Compared to MS2, in AP205 the first β-strand (yellow) is shifted to the C-terminus, although it remains in the same position in 3D. As a result, in AP205, C-and N- termini are the most exposed features on VLPs. In panel (E), crystal structure of AP205 monomer (green) is superimposed with the modeled structure (blue and red). The overall fold of model is approximately correct, except that it lacks C-terminal β-strand. Residues 1-39 (blue) are correctly placed in respect to the sequence, corresponding to the first four β-strands. For the rest of model (red) residues are placed incorrectly according to the sequence and out-of-register errors occur. Notice also that position of N-terminus is relatively well predicted, while C-terminus is in a very different position.