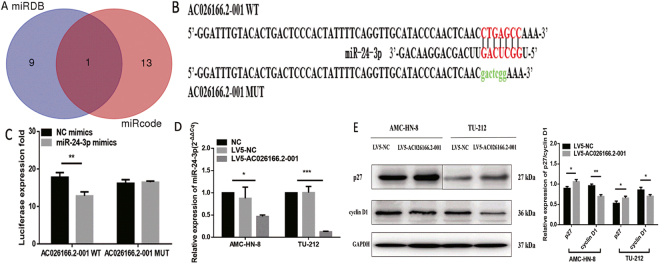
Figure 3.
AC026166.2-001 targets miR-24-3p and promotes p27/Kip1 expression. (A) miRDB: miR-3127-5p, miR-1324, miR-3677-3P, miR-4261, miR-4496, miR-4330, miR-24-3p, miR-219a-2-3p, miR-4727-3p, miR-494-3p. miRcode: miR-190, miR-190a, miR-190b, miR-205, miR-205a, miR-205b, miR-23a, miR-23b, miR-23c, miR-23b-3p, miR-24, miR-24a, miR-24a, miR-24b, miR-24-3p. Both: miR-24-3p. (B) Bioinformatics analysis showed 7 bases in the matched binding sites between AC026166.2-001 and miR-24-3p. As shown above, binding sites or mutated sequence were used for creating firefly luciferase reporter constructs. (C) There are 4 groups (AC026166.2-001 WT/MUT 3′-UTR reporter + miR-24-3p mimics/miR-NC) for the luciferase reporter assay and results demonstrated that miR-24-3p inhibited the AC026166.2-001 WT but not the MUT. (D) After overexpression of AC026166.2-001, miR-24-3p was down-regulated in AMC-HN-8 and TU-212 cell lines. (E) Protein levels of p27, cyclin D1 in AMC-LHN-8 and TU-212 cells transfected with lentiviruses LV5-AC026166.2-001 or LV5-NC determined by Western blot analysis. For details, see Supplementary Figure S1.