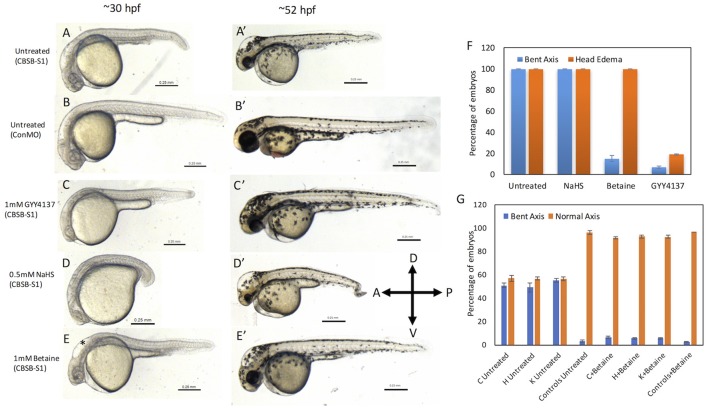
Figure 6.
Rescue of cbsb phenotypes using small molecules at 30 and 52 hpf. All embryos were injected to 2 ng of cbsb-splice1 (CBSB-S1) or control (Con) morpholinos (MOs). Panels (A–E) are 30 hpf phase contrast images, and corresponding (A′-E′) are 52 hpf images. Panels (A,A′) is an untreated embryo injected with CBSB-S1 MO, which shows distinct phenotypes including head edema, and axis defect. Panels (B,B′) are ConMO-injected embryo. Panels (C,C′) are CBSB-S1-injected embryo treated with 1 mM GYY4137, which rescues the head edema and axis defects. Panels (D,D′) are CBSB-S1-injected embryo treated with 0.5 mM NaHS, which does not rescue the phenotype. Panels (E,E′) are CBSB-S1 injected embryo treated with 1 mM betaine, which rescues the axis defect but not the head edema (black asterisk). Panel (F) shows quantification of the rescue of bent axis and head edema phenotypes for CBSB-S1 MO-injected embryos. Y axis shows percentage of embryos with bent axis phenotypes and head edema phenotypes (average of three experiments). Three experiments were performed. X axis categories: Untreated n = 118 embryos, NaHS treated n = 111 embryos, Betaine treated n = 120 embryos and GYY4137 treated n = 121 embryos which are total number of embryos for 3 experiments. ***P < 0.0001 for comparison between untreated and betaine treated for bent axis phenotypes. ***P < 0.0001 for comparison between untreated and GYY4137 treated for both bent axis and head edema phenotypes. Error bars indicate standard error of the mean of three experiments. Panel (G) shows quantification of cbsb CRISPR lines (C, H, and K) for the betaine rescue of axis defects at 5 dpf. For statistical analysis, all embryos with bent axis (irrespective of severity: mild, medium, and severe) were placed into one category bent axis. Y axis shows average of percentage of embryos (with bent axis or normal axis) from three experiments. n = 114 For C Untreated, n = 120 for H Untreated, n = 112 for K Untreated, n = 121 for Controls Untreated, n = 120 for C + Betaine, n = 119 for H + Betaine, n = 124 for K + Betaine and n = 117 for Controls + Betaine, which are total number of embryos from 3 independent experiments. T-test analysis resulted in **P < 0.001 for comparisons between untreated cbsb CRISPR lines and cbsb CRISPR lines treated with betaine. (C vs. C + Betaine, H vs. H + Betaine, K vs. K + Betaine). Differences between control and control + Betaine groups were not statistically significant. Error bars indicate standard error of the mean of three experiments.