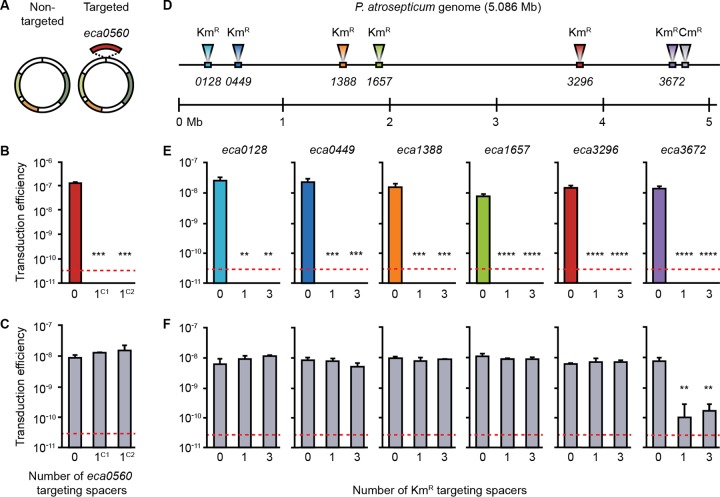
FIG 1 .
CRISPR-Cas can inhibit transduction of plasmids and chromosomal loci. (A) Transducing phage φTE was grown on strains carrying either a control vector (Non-targeted) or a vector with a copy of the eca0560 gene (Targeted). (B and C) The targeted (B) and nontargeted (C) vectors were transduced into strains with either no spacers targeting eca0560 (0; strain ΔHAI2) or one spacer targeting the eca0560 gene in either CRISPR1 (1C1; strain PIM06) or CRISPR2 (1C2; strain PIM17). For additional data from controls, see Fig. S1. (D) Map of the P. atrosepticum genome, indicating the locations of 6 Kmr-marked chromosomal loci and of the secondary Cmr marker in the cas operon in the six strains (strains PCF83 to PCF88). (E and F) The transducing phage, φTE, was grown on these six strains and used to transduce each Kmr marker (E) and Cmr marker (F) into strains either lacking spacers targeting the Kmr gene (0; strain ΔHAI2) or with one (1; strain PIM18) or three (3; strain PIM28) spacers in CRISPR1 targeting the Kmr gene. Data are shown as the mean number of transductants/PFU + standard deviation (SD) (n = 3). The dashed line in each panel represents the limit of detection (one transductant across the replicates). Statistical significance was calculated using one-way analysis of variance (ANOVA) and Dunnett’s multiple-comparison test, comparing strains with targeting spacers to the control with no targeting spacers (**, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001). Additional data from the controls are shown in Fig. S1 and S2.