FIG 2 .
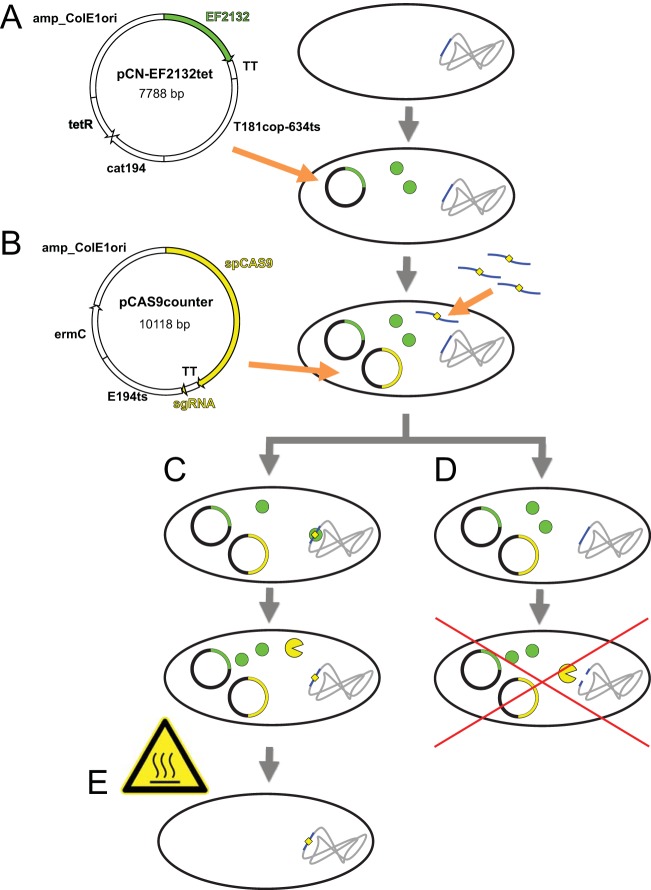
Overview of recombineering and Cas9-mediated counterselection for S. aureus genome engineering. (A) Temperature-sensitive recombineering vector pCN-EF2132tet, encoding recombinase (green circles), is transformed into the strain to be edited. Key elements of the vector are diagrammed and include recombinase EF2132 followed by a transcriptional terminator; high-copy-number temperature-sensitive S. aureus replicon T181cop-634ts; a chloramphenicol resistance gene for selection in S. aureus; and the E. coli ColE1 origin of replication with an ampicillin resistance selectable marker for maintenance in E. coli. (B) Temperature-sensitive counterselection plasmid pCAS9counter is introduced at the time that recombineering is performed. Key elements of the vector are diagrammed and include a synthetic guide RNA (sgRNA) targeted to the genomic site being modified; Cas9 followed by a transcriptional terminator; low-copy-number temperature-sensitive S. aureus replicon E194ts (which is compatible with T181cop-634ts); an erythromycin resistance gene for selection in S. aureus; and the E. coli ColE1 origin of replication with an ampicillin resistance selectable marker for maintenance in E. coli. (C and D) Recombineering is performed using a mutagenic oligonucleotide (blue curved lines) encoding the desired change (yellow diamond). Transformation with these elements leads to two possible outcomes: integration of the mutagenic oligonucleotide is successful and Cas9 (yellow Pac-Man symbol) is unable to cleave the genome (C), or integration of the mutagenic oligonucleotide does not occur and Cas9 introduces a double-stranded break into the host genome, killing unedited cells (D). (E) Brief growth of bacteria at elevated temperatures which are nonpermissive for the plasmid system results in its loss, leaving isogenic, edited cells.