Figure 1. Ineffective Slicing by drAgo2 in Zebrafish Embryos.
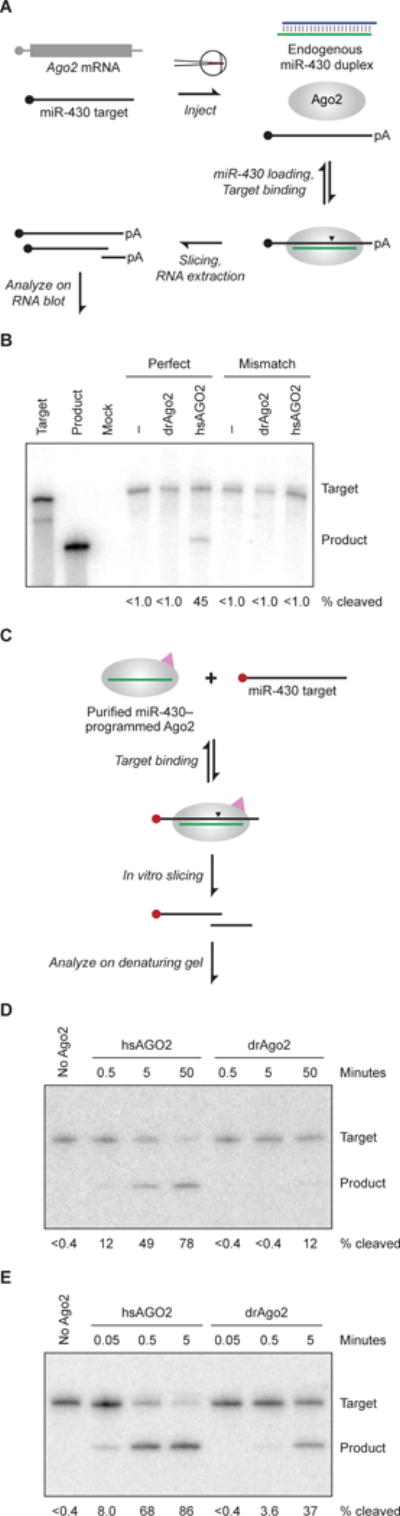
(A) Schematic of the in vivo miR-430–guided slicing assay. Capped target RNA is injected into zebrafish embryos at the one-cell stage, where it becomes polyadenylated (pA) and encounters endogenous miR-430 loaded into either endogenous Ago2 or protein translated from a co-injected Ago2 mRNA. To the extent that Ago2 is capable, it slices the target at the site indicted (arrowhead). Injected embryos develop until 4 hpf, at which point RNA is extracted and remaining target and sliced product are resolved and detected on an RNA blot.
(B) Slicing activities of zebrafish and human proteins in zebrafish embryos. Shown is an RNA blot probing for an injected target with or without a perfectly complementary miR-430 site (perfect and mismatch, respectively) and with or without co-injection of mRNA for the indicated protein. The left two lanes are uninjected standards representing the full-length target and sliced product. Mock is an uninjected negative control. Below each lane is the percentage of total signal (target plus product bands) represented by product (% cleaved).
(C) Schematic of the in vitro slicing assay, which uses Ago2 that was affinity-purified based on both its association with a specific miRNA (miR-430 in this example, green line) and its 3X-FLAG tag (pink triangle). After binding to the purified complex, cap-labeled (red filled circle) target is sliced at the site indicated (arrowhead), and labeled product is resolved from target on a denaturing gel.
(D) In vitro slicing activities of human and zebrafish Ago2 proteins programed with miR-430. Shown is a denaturing gel resolving cap-labeled target and product after incubation of limiting target (0.1 nM) with the indicated Ago2 protein (1.0 nM) for the indicated time. Below each lane is the percentage of target converted to product, reporting the mean from two experiments.
(E) In vitro slicing activities of human and zebrafish Ago2 proteins programmed with miR-1. Below each lane is the percentage of target converted to product. Otherwise, this panel is as in (D).
See also Figure S1.
