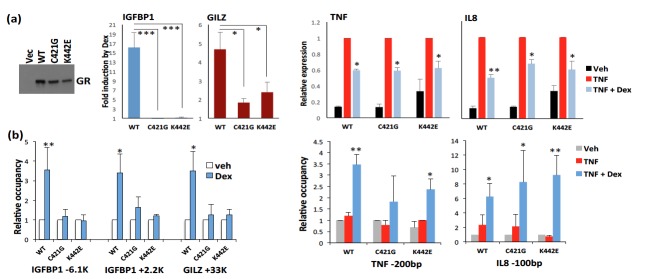
Author response image 1. GR DNA-binding mutants are competent for repression at NF-κB GR-tethering sites.
U2OS cells were transfected with hGR (WT, C421G or K442E)-expressing or empty EF.CMV.RFP vector (a, left) using Lipofectamine 3000 (ThermoFisher) and treated the next day with vehicle (Veh), 100 nM Dex, 5 ng/ml TNF or TNF+Dex, as indicated. (a) Gene expression of GR-activated IGFBP1 and GILZ (middle) or GR-repressed TNF and IL8 (right) following 2 h treatment was assessed by RT-qPCR with b-Actin as housekeeping control. IGFBP1 (n=5) and GILZ (n=4) fold induction by Dex is expressed over transcript level in veh-treated cells (=1). TNF and IL8 expression (n=3 each) is calculated relative to the transcript level in TNF-treated cells (=1). (b) ChIP with polyclonal antiGR Abs (PA1-511A ThermoFisher) was performed following 2-h Dex (left) or 1-h TNF or TNF+Dex (right) treatment, and GR occupancy was assessed at GREs and NF-κB sites at indicated genomic locations with signals at 28S ribosomal gene as normalization control (n=3). Occupancy is expressed over baseline in veh-treated cells (=1). Error bars are SEM; 2-tailed Student’s t-test.