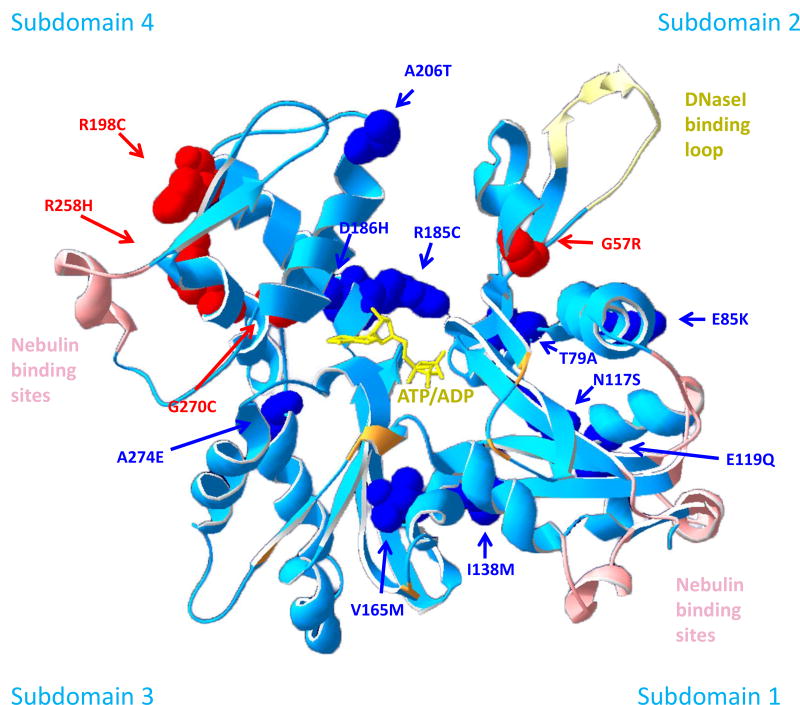
Figure 1.
Molecular model of an actin monomer highlighting the position of mutated residues analyzed in this study (the molecular model of alpha actin was based on a 2.8 Å atomic structure of rabbit skeletal muscle actin (RCSB Protein Data Bank 1ATN,36). The ribbon structure showing side chains affected by ACTA1 assessed in this study was created with Swiss-PDB Viewer v4.1.037). Human alpha skeletal actin is a globular molecule consisting of 2 domains (4 subdomains) which are connected by a “hinge” region (ribbon structure shown in light blue). A cleft is located between the two domains containing a divalent cation and ATP/ADP (yellow). Actin polymerizes into two twisted strands where each actin molecule interacts with four neighboring monomers. The DNase1 binding loop (pale yellow) is involved in actin-actin binding by forming a lock-in-key interaction between neighboring actin molecules. Details on further residues involved in actin-actin interactions can be found in53. Actin binds to a number of proteins which are crucial for its function. Important binding partners include nebulin (amino acids involved in binding are 98–103, 127–132, 361–368, 228–238, 4–8, 350–358, shown in light pink53), tropomyosin (Asp27, Arg30, Arg149, Lys328, Lys329, Lys330, Pro335 and Glu336,56, shown in orange) and myosin (via amino acid 2–6, 26–27, 101–102, 146, 334, 335, 343, 351, 35453). The 14 actin residues affected by the mutations we investigated in this study are highlighted in red and blue depending on their effect on the Ca2+ sensitivity of force generation (red = reduced Ca2+ sensitivity; dark blue = Ca2+ sensitivity comparable to controls). Three of four residues reducing Ca2+ sensitivity are located within subdomain 4.