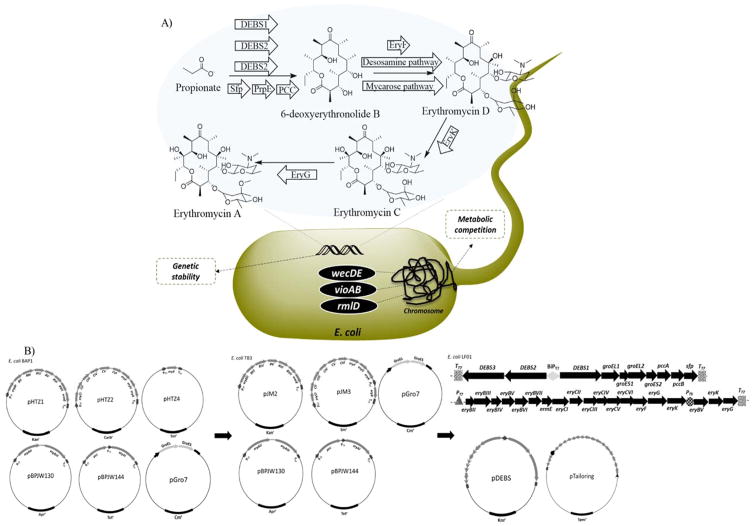
Figure 1.
E. coli heterologous erythromycin biosynthesis and plasmid re-design. A) The heterologous erythromycin pathway established in E. coli featuring the DEBS1, 2, and 3 polyketide synthases, Sfp (required for polyketide synthase posttranslational modification), PrpE (a propionyl-CoA synthetase native to E. coli), PCC (a propionyl-CoA carboxylase), EryF (hydroxylase), the desosamine and mycarose deoxysugar pathways, EryK (hydoroxylase), and EryG (methylase). Engineering steps were taken to improve metabolic conversion and genetic stability. B) The left5, middle12, and right (this study) successive plasmid designs to address stability. PT7: T7 promoter; BiPT7: bi-directional T7 promoter; TT7: T7 terminator sequence; PT5: T5 promoter.