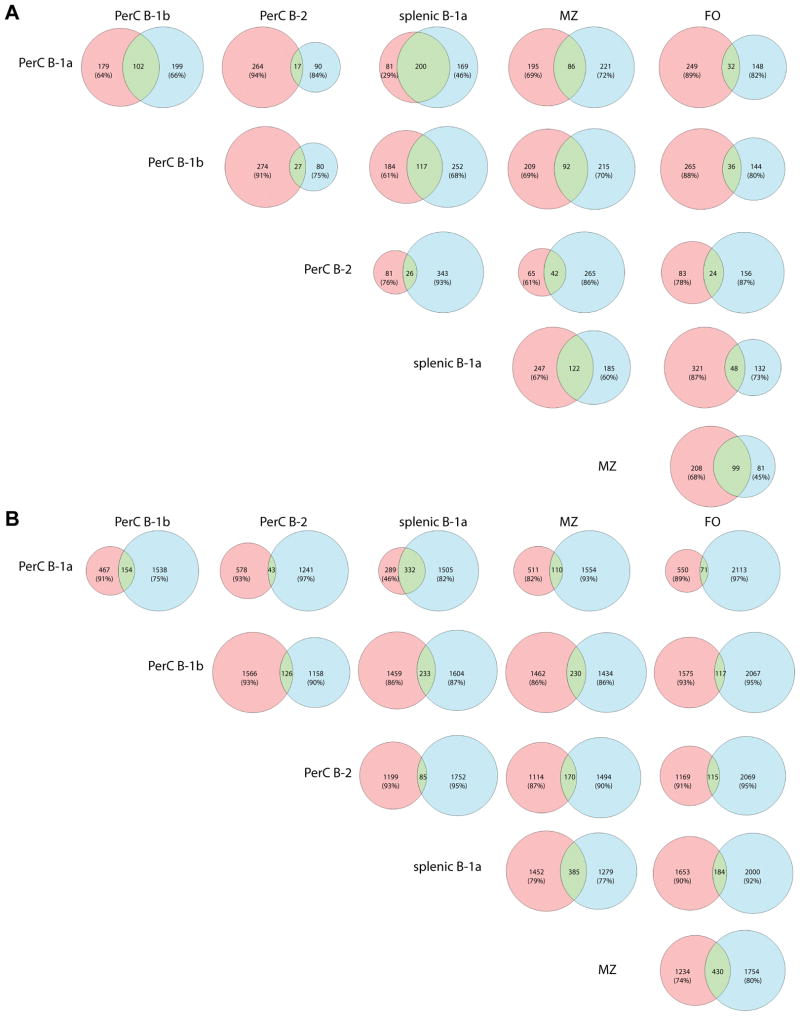
Figure 9. Overlap of N− and N+ CDR3 repertoires.
Applying the same rules as in Figure 8, we separately compared sequences with and without N additions. (A) First, only N− sequences were analyzed. Most notably there was marked overlap between the N− repertoires of peritoneal and splenic B-1a cells. These cell subsets shared more N− sequences than there are unique sequences in each of the populations. The MZ subset shared a significant number of N− sequences with all other subsets (14–40% of its N− sequences). B-2 cells have a low absolute number of N− sequences – however, 39.2% of these B-2 N− sequences also occur in MZ cells, which is the closest relationship the B-2 repertoire has with any other cell type. FO cells shared a substantial amount of their N− sequences with MZ cells, but notably only few with B-2 cells. (B) Analyzing only N+ sequences, there again was a large number of sequences shared between peritoneal and splenic B-1a cells, but the splenic B-1a subset included many more N+ sequences than the peritoneal. There was also marked overlap between splenic B-1a and MZ cells – in absolute numbers, they shared more N+ sequences than peritoneal and splenic B-1a cells. MZ cells also shared a significant portion of their transcripts with FO cells, which was the biggest absolute number of shared N+ sequences between any subset. Peritoneal B-2 cells proved to be an isolated subset, having the most overlap with MZ cells, but still at a low quantity of only 13.2% of their N+ repertoire.