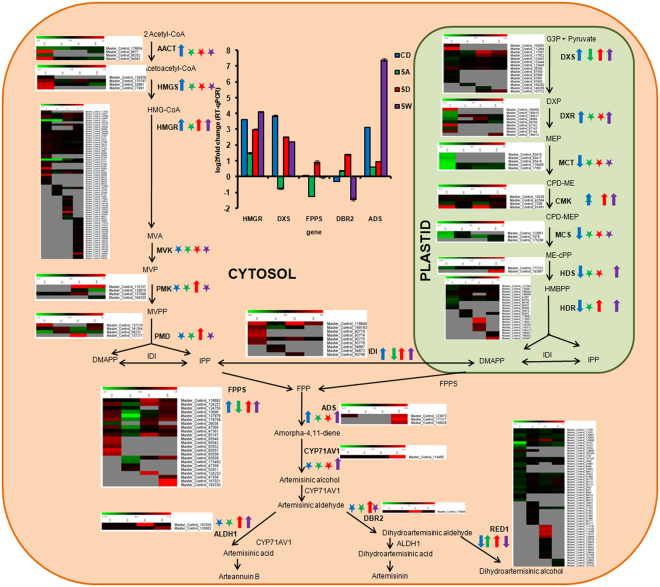
Figure 7.
Differentially expressed artemisinin and other terpenoid biosynthetic pathway genes of A. annua during abiotic stresses. Expression patterns of artemisinin and other terpenoid biosynthetic pathway genes were represented based on average log2fold change values for the genes showing differential expression (calculated from transcripts with log2fold change >+1 or <−1). Upward arrows indicate up-regulation, downward arrows indicate down regulation and stars indicate neutral regulation or genes not identified. Graph represents validation of the expression pattern of five differentially expressed genes by RT-qPCR (data was analyzed by three independent repeats and standard deviations were shown with error bars). Color coding: blue for cold, green for salt, red for drought and purple for water-logging stress. Heat maps demonstrate the log2fold change values of all differentially expressed transcripts corresponding to a particular gene. Abbreviations used: CD,cold; SA,salinity; SD,drought; SW, water-logging; HMG-CoA, Hydroxymethylglutaryl-coenzyme A; MVA, mevalonic acid; MVP, 5-phosphomevalonate; MVPP, 5-diphosphomevalonate; DMAPP, dimethylallyl diphosphate; IPP, isopentenyl diphosphate; FPP, farnesyl diphosphate; AACT, acetoacetyl-coenzyme A thiolase; HMGS, 3-hydroxy-3-methyl-glutaryl coenzyme A synthase; HMGR, 3-hydroxy-3-methyl-glutaryl coenzyme A reductase; MVK, mevalonate kinase; PMK, phosphomevalonate kinase; PMD, diphosphomevalonate decarboxylase; IDI, isopentenyl diphosphate isomerase; FPPS, farnesyl diphosphate synthase; G3P, glyceraldehyde-3-phosphate; DXP, 1-deoxy-d-xylulose-5-phosphate; MEP, 2-C-methyl-d-erythritol-4-phosphate; CDP-ME, 4-(cytidine 5′-diphospho)-2-C-methyl-d-erythritol; CDP-MEP, 2-phospho-4-(cytidine 5′-dipospho)-2-C-methyl-d-erythritol; ME-cPP, 2-C-methyl-d-erythritol-2,4-cyclodiposphate; HBMPP, hydroxymethylbutenyl-4-diphosphate; DXS, 1-deoxy-d-xylulose-5-phosphate synthase; DXR, 1-deoxy-dxylulose-5-phosphate reductoisomerase; MCT, 2-C-methyl-d-erythritol-4-(cytidyl-5-diphosphate) transferase; CMK, 4-cytidine 5′-diphospho-2-C-methyl-d-erythritol kinase; MCS, 2-C-methyl-d-erythritol-2,4-cyclodiphosphate synthase; HDS, hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate synthase; HDR, hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate reductase, ADS, amorpha-4,11-diene synthase; ALDH1, aldehyde dehydrogenase 1; CYP71AV1, amorphadiene-12-hydroxylase; DBR2, artemisinic aldehyde reductase17.