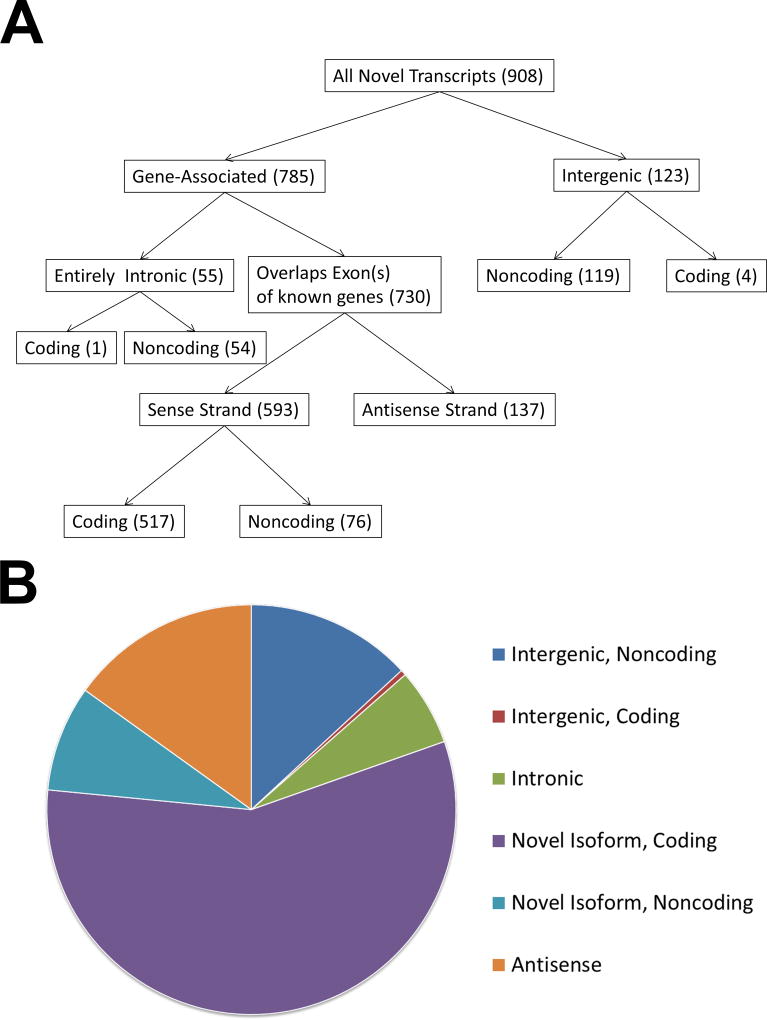
Figure 1.
A flow chart (A) and pie chart (B) depicting and summarizing the filtering categories used to classify novel transcripts identified in this study. The categories for intronic, novel isoforms, antisense, and intergenic transcripts were defined via a CuffCompare annotation using the Comprehensive Gencode Resease 25 annotation (hg19) reference transcriptome. Coding potential of novel transcripts was predicted using CPAT.