Figure 2.
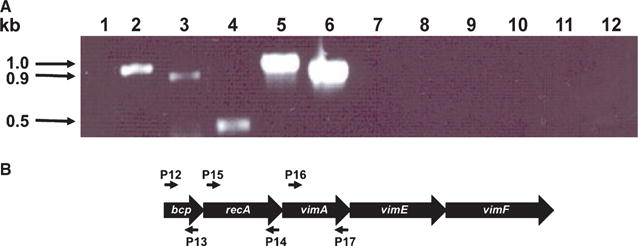
(A) Reverse transcription–polymerase chain reaction (RT-PCR) analysis of RNA extracted from Porphyromonas gingivalis W83 and FLL302. Total RNA was extracted from FLL302 (lanes 1–3 and 7–9) and W83 (lanes 4–6 and 10–12) grown to mid log-phase (OD600, 0.6) and subjected to RT-PCR. Porphyromonas gingivalis FLL302: lane 1, primers (P1 and P2, Table 1) plus reverse transcriptase; lane 2, primers (P16 and P17, Table 1) plus reverse transcriptase; lane 3, primers (P14 and P15, Table 1) plus reverse transcriptase; P. gingivalis W83: lane 4, primers (P1 and P2, Table 1) plus reverse transcriptase; lane 5, primers (P16 and P17, Table 1) plus reverse transcriptase; lane 6, primers (P14 and P15, Table 1) plus reverse transcriptase; lanes 7–12, corresponding primer sets for the bcp, recA and vimA genes minus reverse transcriptase (negative controls). All lanes contain 5 μl of the amplified mixture. (B) Location of the primers used in RT-PCR analysis.
