Fig 4. Structural analysis of escape mutations.
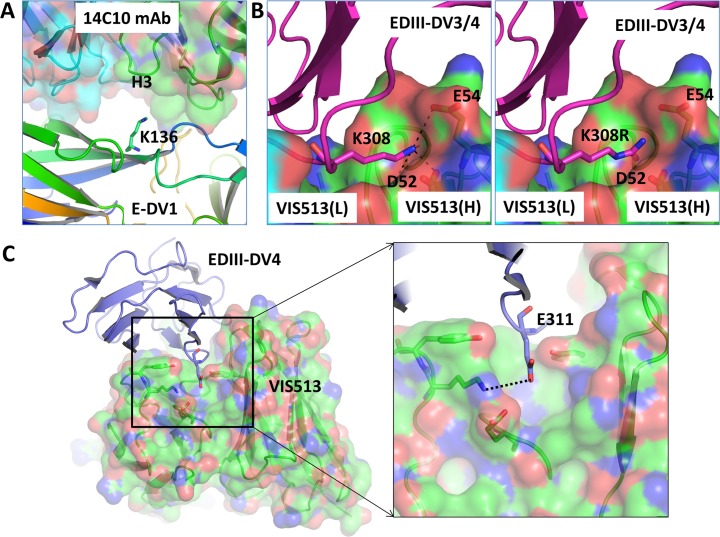
A. Molecular interactions of E-DENV1 protein (shown in rainbow color cartoon) are shown with the 14c10 antibody (shown in transparent surface and cartoon diagram with heavy and light chains shown in green and cyan color, respectively). In the complex, the Lys136 residue of the E-DENV1 protein is found in the vicinity of the HCDR3 loop of the mAb. Mutation of Lys136 to a Glu is expected to cause loss of mAb binding due to the change in charge of the residue. B. Molecular interactions of Lys308/310 of EDIII-DENV3/4 protein (shown in magenta cartoon) are shown with VIS513 (shown in transparent surface and cartoon diagram with heavy and light chains shown in green and cyan color, respectively). The Lys308 of E-DENV3 or Lys310 of E-DENV4 residues are the same and it is found to form hydrogen bonding and salt bridges with the VIS513 heavy chain residues Asp52 and Glu54 (left panel). The right panel shows the modeling of mutation Lys308Arg. C. Modeling of position E311 of E-DENV4 region found to be mutated in virus from mouse samples. A hydrogen bond with the antibody molecule is shown with dotted line.