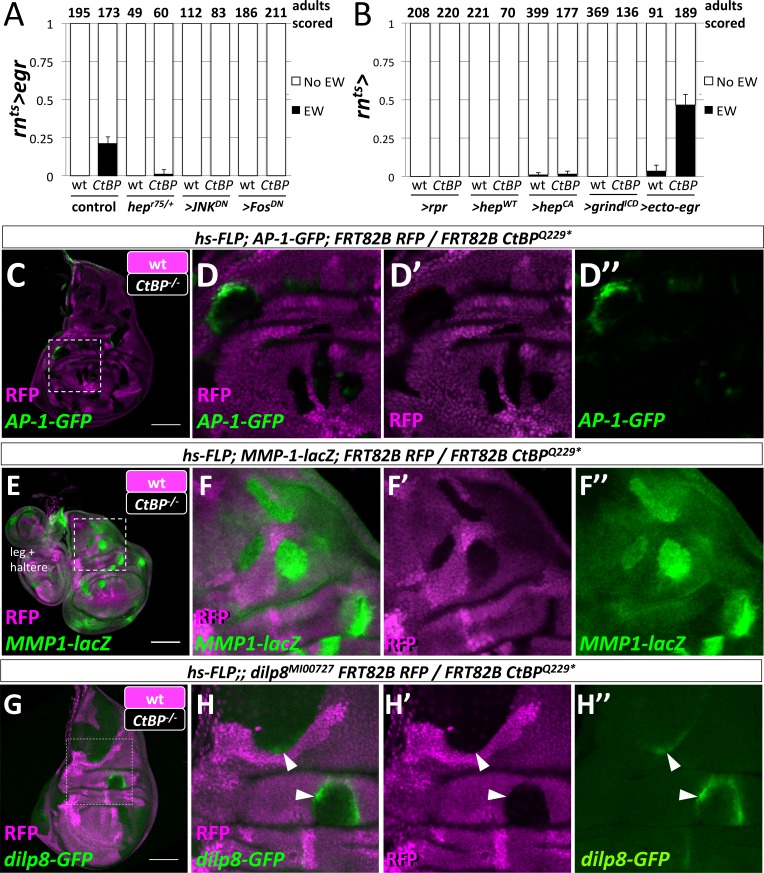
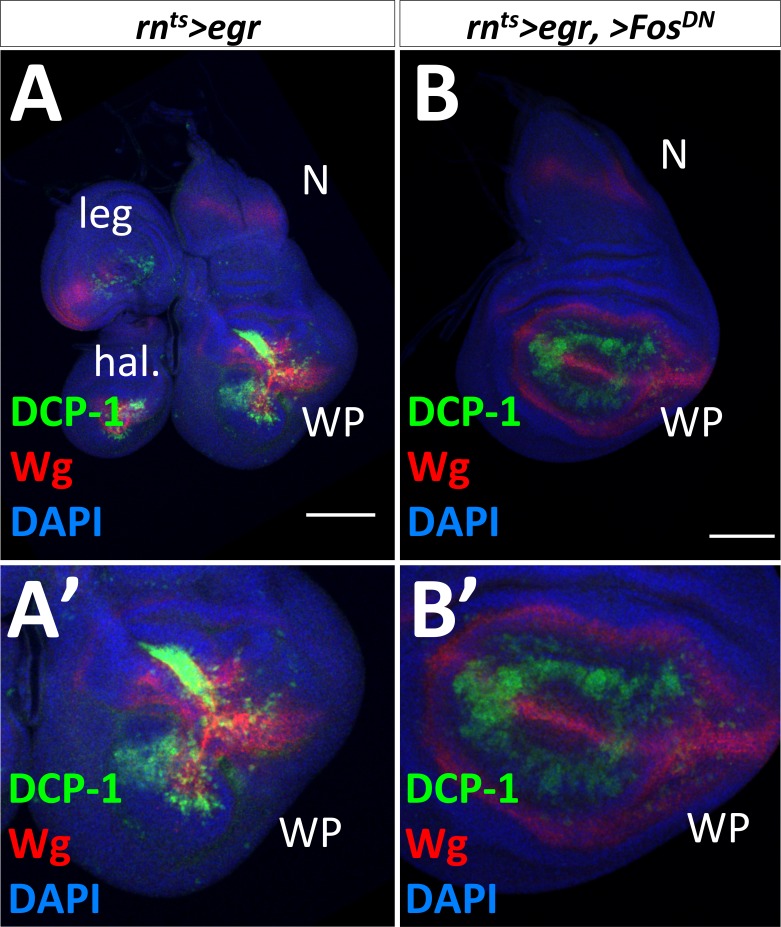
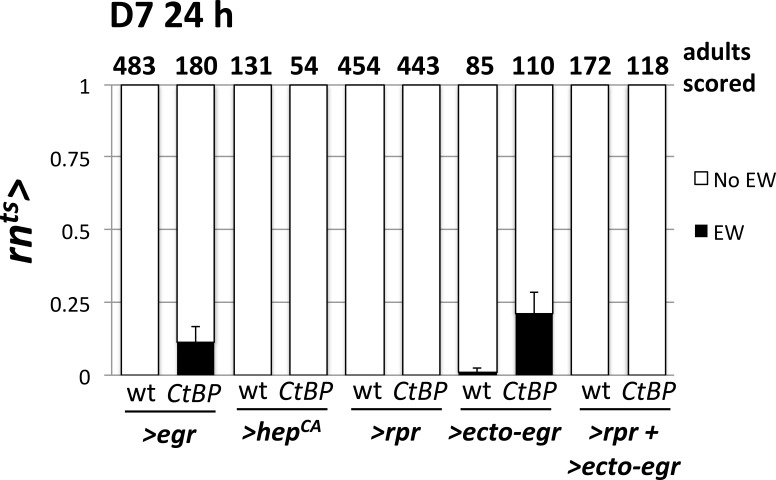
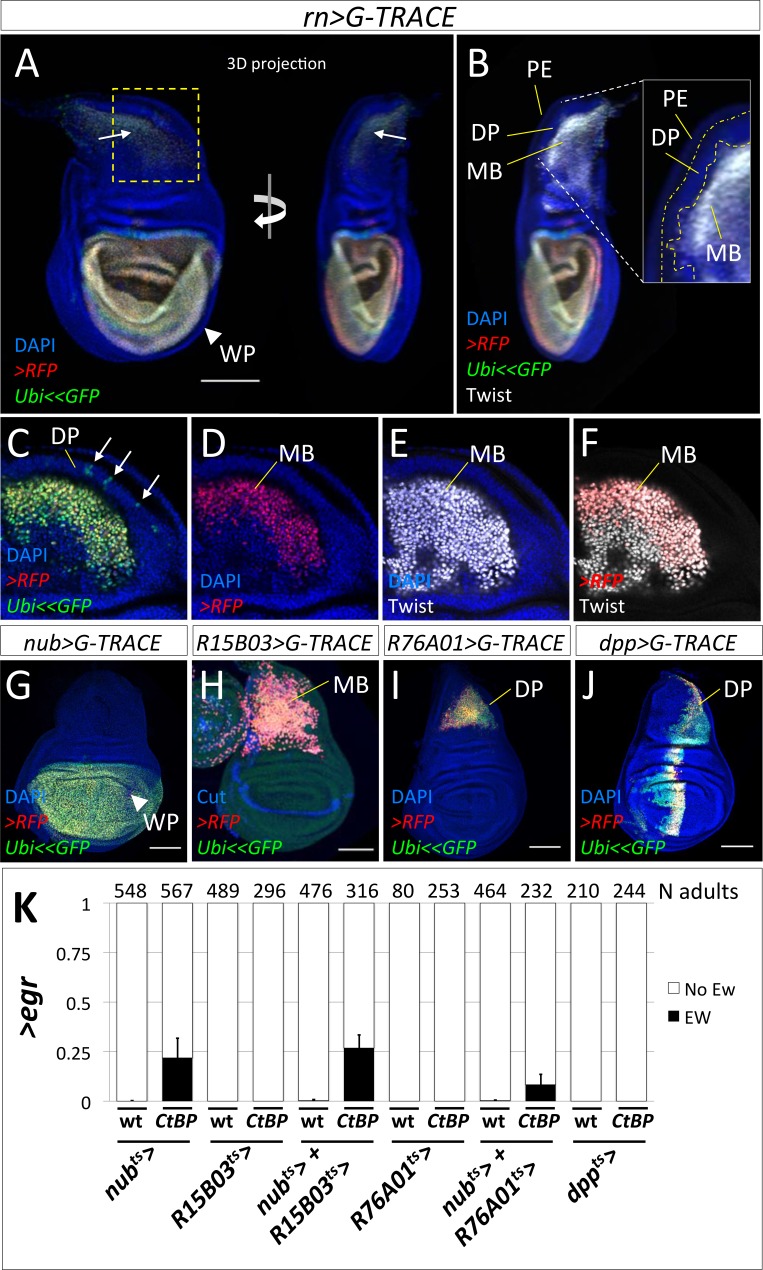
Figure 5. Egr activation of the JNK pathway is required for the formation of ectopic wings.
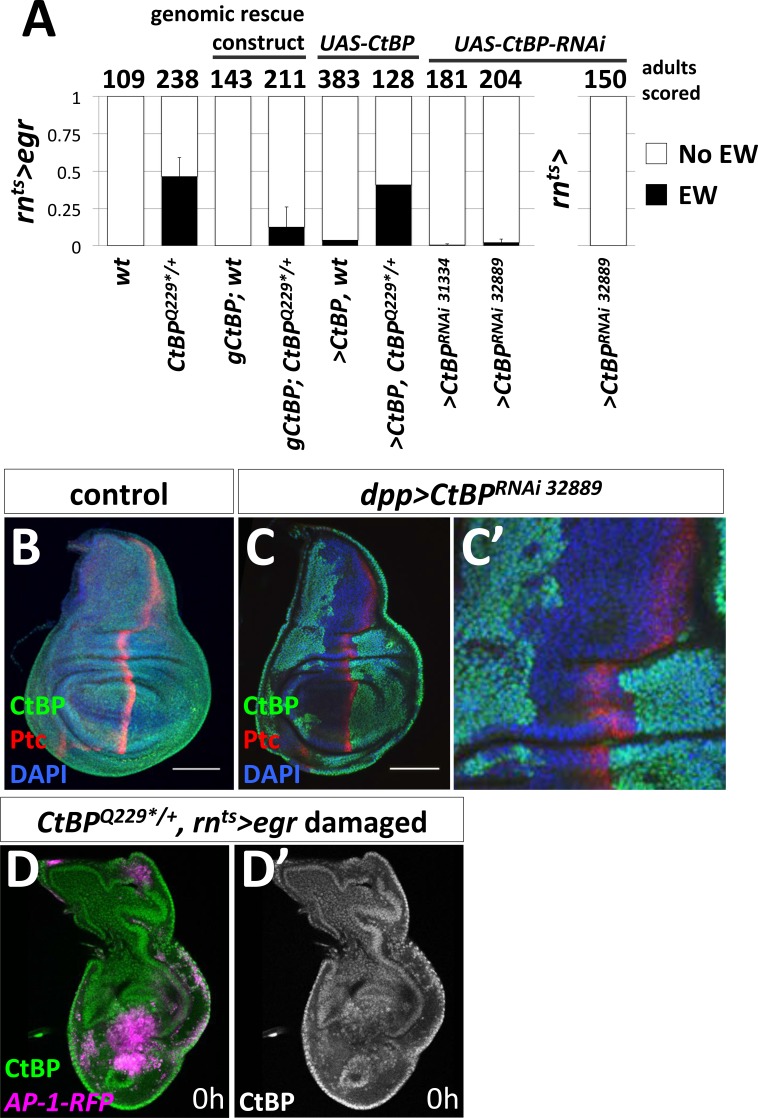
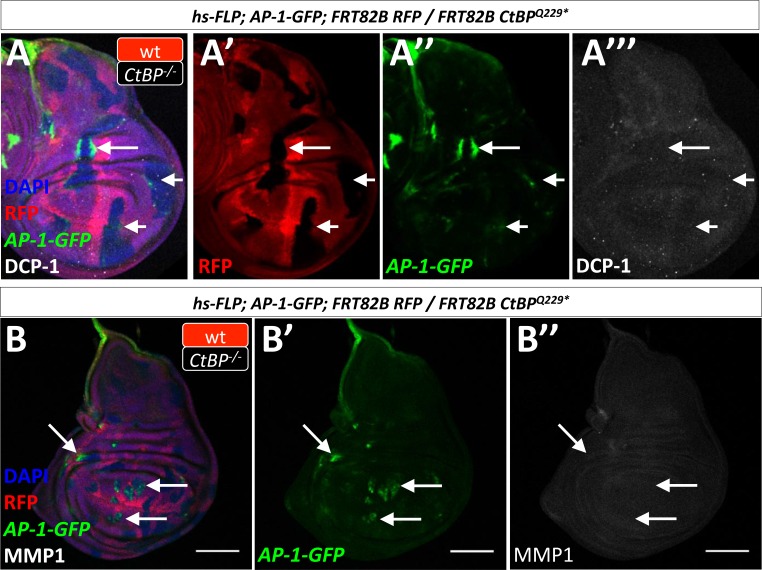
(A) Frequency of ectopic wings (EW) following rnts>egr damage in wild type or CtBP334Δ4/+ (CtBP334Δ4 was generated via CRISPR/Cas9 on the same chromosome that contains rnts>egr) in listed genotypes: (1) hepr75/+, (2) co-expression of UAS-JNKDN and (3) co-expression of UAS-FosDN. (B) Frequency of ectopic wings (EW) following rnts> expression of listed transgenes (UAS-rpr, UAS-hepwt, UAS-hepCA, UAS-grindICD, UAS-ecto-egr) in wild type and CtBP-/+ genetic background. Temperature shifts for experiments shown in (A) and (B) were done on day 7 AEL for 40 hr. (C–H) Wing discs with homozygous mutant clones of FRT82B CtBPQ229* generated by mitotic recombination with the transcriptional reporters for AP-1-GFP (C, D), MMP1-lacZ (E, F) and dilp8-GFP (G, H).