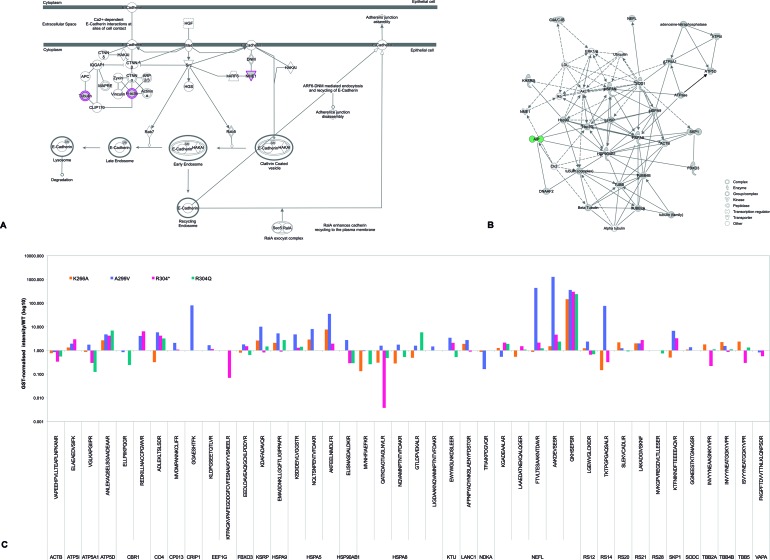
Figure 1. AIP candidate interacting partners: signaling pathways and differential peptide repertoires in pull-down experiments.
(A) The top signaling pathway reported by Ingenuity Pathway Analysis when analyzing the 30 candidate interacting partners identified for WT AIP (as listed in Table 3) was “Remodeling of the epithelial adherens junction” (P < 0.0001). This pathway included five candidate partners (ACTB, NME1, TUBB, TUBB2A, and tubulin beta-4B chain [TUBB4B]) out of the total of 68 proteins reported for such pathway by the platform used (overlap: 7.4%). Proteins in this pathway represent multiprotein complexes, with cadherins as central components, mediating cell-cell adhesion and intercellular communication, and regulating cell shape and polarity. Other top canonical pathways reported by this analysis were “Regulation of eIF4 and p70S6K signaling” (5/157 proteins [3.2%], P < 0.0001), “EIF2 signaling” (5/194 [2.6%], P < 0.0001), “mTOR signaling” (5/199 [2.5%], P < 0.0001) and “Protein ubiquitination pathway” (5/255 [2.5%], P < 0.0001). Proteins marked in pink are among the AIP candidate interacting partners identified by our pull-down experiments. F-actin refers to “filamentous actin” (i.e. a polymer of actin molecules, including ACTB among other isoforms) which forms part of the cytoskeleton. (B) Nineteen of the AIP WT candidate partners were grouped together in a single network, either due to functional relationships or direct binding. (C) Schematic representation of the pull-down data presented in Supplementary Table 2 (in a logarithmic scale for easy overview) including only proteins present in the AIP WT experiment. Proteins whose peptides were underrepresented in the AIP variant protein pull-down experiments were interpreted as impaired or lost interactions.