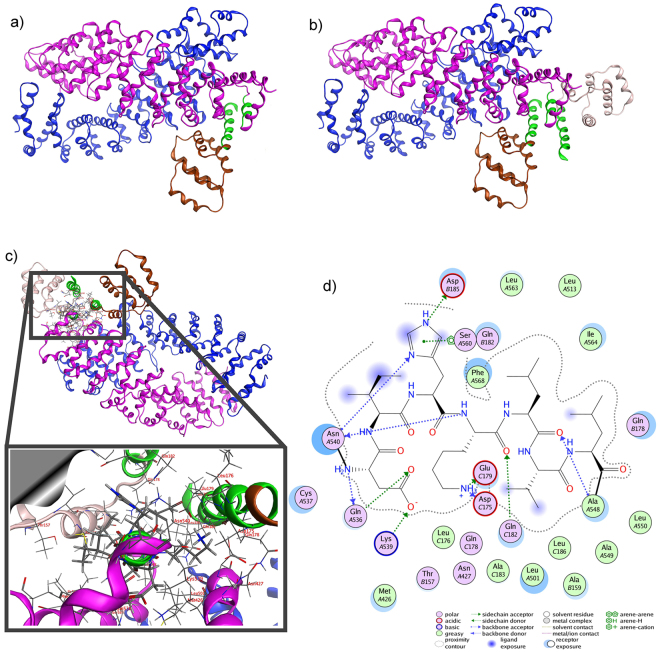
Figure 5.
Molecular docking of LRRK2 ARM to FADD-DD. The model of dimeric LRRK2 ARM was used to dock the FADD DD crystal structure. The docking was performed iteratively placing each FADD DD structure onto the LRRK2 ARM model in sequence. (a) The FADD DD crystal structure (brown ribbon) was docked onto the LRRK2 dimer model (shown in blue and magenta ribbon, as before). (b) A second copy of the FADD DD crystal structure (silver ribbon) was docked to the molecular complex established in the previous step (a). The interaction was achieved via a network of antiparallel α-helical bundles (shown in green ribbon representation). (c) The network of molecular interactions between the specific residues involved is highlighted (inset). (d) The association between LRRK2 and FADD DD is stabilized via a mixture of ionic/hydrogen bonds, π-stacking and hydrophobic interactions. The amino acid notations are as follows: A, refers to the LRRK2 ARM sequence; and B, C refer to each FADD DD molecule.