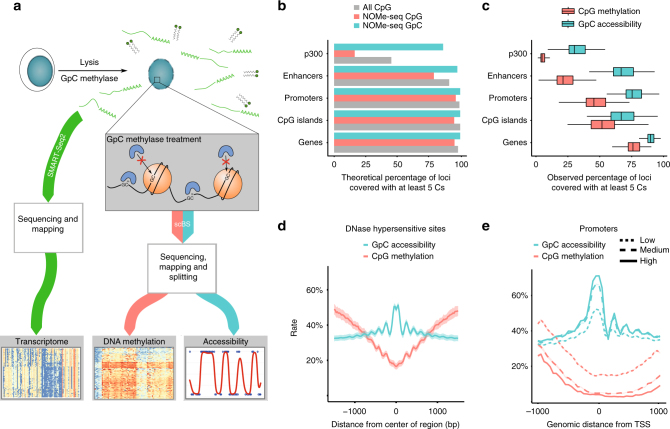
Fig. 1.
scNMT-seq overview and genome-wide coverage. a Protocol overview. Single-cells are lysed and accessible DNA is labelled using GpC methyltransferase. RNA is then separated and sequenced using Smart-seq2, whilst DNA undergoes scBS-seq library preparation and sequencing. Methylation and chromatin accessibility data are separated bioinformatically. b Theoretical maximum CpG coverage of genomic contexts with known regulatory roles. Shown is the proportion of loci in different contexts that contain at least 5 cytosines. ‘All CpG’ considers any C-G dinucleotides (e.g., as in scBS-seq), ‘NOMe-seq CpG’ considers A–C–G and T–C–G trinucleotides and ‘NOMe-seq GpC’ considers G–C–A, G–C–C and G–C–T trinucleotides. c Empirical coverage in 61 mouse ES cells considering the same contexts as in b. Shown is the coverage distribution across cells after QC; box plots show median coverage and the first and third quartile, whiskers show 1.5 × the interquartile range above and below the box. d CpG methylation and GpC accessibility profiles at published DNase hypersensitive sites19. The profiles were computed as a running average in 50 bp windows. Shading denotes standard deviation across cells. e CpG methylation and GpC accessibility profiles at gene promoters. Promoters are stratified by average expression level of the corresponding gene (log normalised counts less than 2 (low), between 2 and 6 (medium) and higher than 6 (high). The profile is generated by computing a running average in 50 bp windows