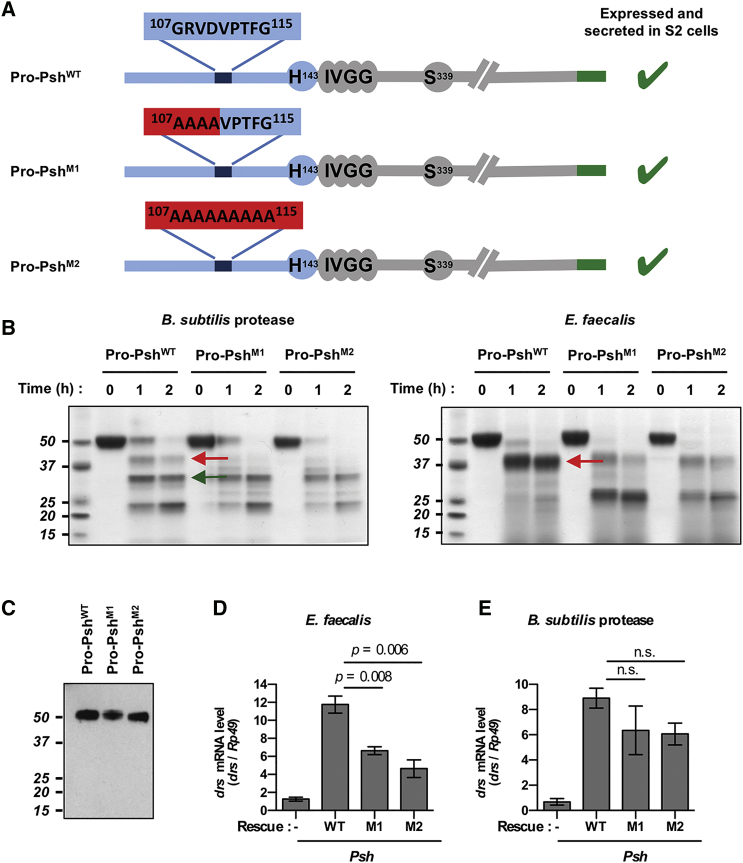
Figure 4.
Psh Inactivation by Mutations in the Bait Region
(A) Structure of pro-Psh mutants with partial (pro-PshM1) or total substitution (pro-PshM2) of the bait region by alanyl residues.
(B) Purified mutant or wild-type rpro-Psh (0.2 μg/μL) was incubated for 1 or 2 hr at 29°C with cell-free E. faecalis culture medium supernatant or with purified B. subtilis protease (1 nM). Following electrophoresis, hydrolysis products were visualized by Coomassie blue staining. Red arrows indicate fragments resulting from hydrolysis in the bait region and the green arrow indicates the active catalytic domain.
(C–D) Wild-type pro-Psh, pro-PshM1, and pro-PshM2 were expressed under the control of the fat body Yolk driver in psh1 mutant flies.
(C) Secretion of wild-type and mutant pro-Psh in the blood was determined by western blot using an anti-6HisTag antibody.
(D and E) drs mRNA levels were monitored by qPCR in psh1 mutant flies (psh1; yolk-GAL4/+) expressing the wild-type rpo-Psh (WT; psh1; yolk-GAL4/UAS-pro-Psh), the M1 mutant (M1; psh1; yolk-GAL4/UAS-pro-PshM1), and the M2 mutant (M2; psh1; yolk-GAL4/UAS-proPshM2) for 24 hr at 25°C following E. faecalis challenge (OD600 = 1) (D) or 16 hr at 25°C following B. subtilis protease injection (E).
Data represent means ± SEs of three independent experiments, each containing three groups of ten flies (five males and five females). p values obtained from Student’s t test are indicated on the graphs.