FIGURE 3.
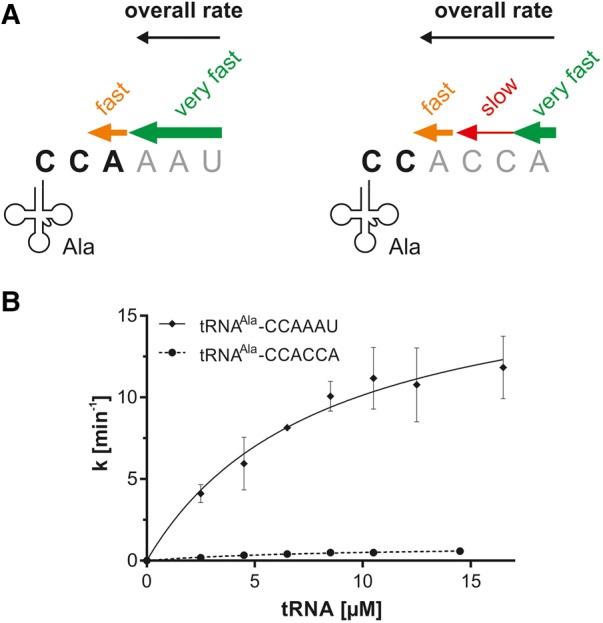
RNase T kinetics on tRNA substrates with 3′-overhangs of similar length. (A) Our initial experiments showed that tRNAs are degraded to different extents, depending on the sequence of the 3′-extension (reaction velocity is indicated by size and color of the individual arrows; experimental data are not shown). For the kinetic analysis, main reaction products able to reenter the functional tRNA pool were taken into account (3′-end in bold characters). tRNA–CCAAAU with the natural trailer (left) is rapidly degraded to the CCA end as the main reaction product (green arrow) that was analyzed in the kinetic analysis. The subsequent 3′-end turnover (removal of the terminal A residue, orange arrow) is slower and therefore was not considered. Individual bases of the degradation tag CCACCA are removed at different reaction velocities (right). Although the terminal A residue of the CCACCA sequence is also removed very fast (green arrow), degradation of the two following C residues is very slow (red arrow) and corresponds to the rate-limiting step of the reaction. The subsequent 3′-end turnover (removal of the terminal A residue of the resulting CCA end, orange arrow) is then removed very efficiently. Hence, for this tRNA, tRNA–CC represents the resulting main reaction product for the kinetic analysis. (B) Kinetic analysis of tRNAAla–CCAAAU trailer removal by RNase T. The CCACCA tag is nearly not converted. k is the reaction rate normalized to the total enzyme concentration. Data are means ± SD; n = 3–4. A different scale of the kinetics of the CCACCA removal is displayed in Supplemental Figure S2.
