Abstract
Many studies have examined the associations between paraoxonase‐1 (PON1) genetic polymorphisms (Q192R, rs662 and L55M, rs854560) and the susceptibility to type 2 diabetes mellitus (T2DM) across different ethnic populations. However, the evidence for the associations remains inconclusive. In this study, we performed a meta‐analysis to clarify the association of the two PON1 variants with T2DM risk. We carried out a systematic search of PubMed, Embase, CNKI and Wanfang databases for studies published before June 2017. The pooled odds ratios (ORs) for the association and their corresponding 95% confidence intervals (CIs) were calculated by a random‐ or fixed‐effect model. A total of 50 eligible studies, including 34 and 16 studies were identified for the PON1 Q192R (rs662) and L55M (rs854560) polymorphism, respectively. As for the PON1 Q192R polymorphism, the 192R allele was a susceptible factor of T2DM in the South or East Asian population (OR > 1, P < 0.05) but represented a protective factor of T2DM in European population (OR = 0.66, 95% CI = 0.45–0.98) under a heterozygous genetic model. With regard to the PON1 L55M polymorphism, significant protective effects of the 55M allele on T2DM under the heterozygous (OR = 0.77, 95% CI = 0.61–0.97) and dominant (OR = 0.80, 95% CI = 0.65–0.99) genetic models were found in the European population, while no significant associations in the Asian populations under all genetic models (P > 0.05). In summary, by a comprehensive meta‐analysis, our results firmly indicated that distinct effects of PON1 genetic polymorphisms existed in the risk of T2DM across different ethnic backgrounds.
Keywords: type 2 diabetes mellitus, susceptibility, paraoxonase‐1, polymorphism, ethnic difference
Introduction
The rise of diabetes prevalence poses one of the important challenges to global health. It is estimated that approximately 422 million adults were diagnosed with the disease in 2014 worldwide 1. Diabetes is one of the main causes of cardiovascular disease, blindness and kidney failure and is the sixth leading driver of disability 2. Therefore, the prevention and control of diabetes are growing up to be an ever‐increasing global health priority 3. Type 2 diabetes mellitus (T2DM) comprises the majority of cases of diabetes around the world. T2DM is a metabolic disorder of multifactorial aetiology involving many environmental factors and genetic variants 4, 5.
Human paraoxonase‐1 (PON1) is a calcium‐dependent 45‐kD glycoprotein composed of 355 amino acids. The esterase is synthesized mainly by the liver and secreted into the circulation where it associates with high‐density lipoprotein (HDL) and assists in the antioxidant effect of preventing oxidation of low‐density lipoprotein (LDL). PON1 in human beings is encoded by the PON1 gene which maps to the long arm of chromosome 7 (q21‐22). It has been observed that serum PON1 activity has an important role in susceptibility and progression of T2DM 6, 7.
Single nucleotide polymorphisms (SNPs) in the PON1 gene can significantly account for the catalytic ability of the enzyme. A missense SNP at position 192 (glycine (Q) to arginine (R) substitution) (rs662) is an important determinant of the PON1 activity 8. Although the R‐alloenzyme is more active towards some substrates, for example paraoxon, other substrates such as diazoxon and sarin are hydrolysed more rapidly by the Q‐alloenzyme 9. In addition, the PON1 Q192R polymorphism was the major determinant of individual variation in the ability of HDL in protecting LDL against lipid peroxidation. For example, the Q‐alloenzyme confers least ability 10. Another SNP in the coding region causes a leucine (L) to methionine (M) substitution at position 55 (rs854560), which may also affect the PON1 activity and levels 11.
As Ikeda et al. first found that serum PON activity was significantly decreased in the patients with T2DM 12, a large number of studies have been conducted over the last two decades to investigate the association of Q192R (rs662) and L55M (rs854560) polymorphism in PON1 gene with susceptibility to T2DM. However, the previously published results remain controversial. Hence, to firmly elucidate the association between PON1 genetic polymorphisms (Q192R, rs662 and L55M, rs854560) and the risk of T2DM, we conducted a systematic review and meta‐analysis of data from 50 studies and also established the association according to the ethnicity.
Materials and methods
Search strategy and inclusion criteria
A systematic search was conducted in the electronic databases PubMed, Embase, China National Knowledge Infrastructure (CNKI) and Wanfang Data, and all relevant articles were published in English or Chinese from their starting dates to June 2017. The search strategy used the following keywords relating to the paraoxonase‐1 gene (‘paraoxonase‐1′, ‘PON1′) or variations (e.g. ‘mutation’, ‘polymorphism’, ‘single nucleotide polymorphism’, ‘SNP’, ‘variant’, ‘variation’) in combination with TD2M (e.g. ‘Diabetes Mellitus, Type 2′, ‘Noninsulin‐Dependent Diabetes Mellitus’, ‘Type 2 Diabetes’, ‘Diabetes Mellitus, Noninsulin‐Dependent’). We supplemented this search by reviewing the cited references for all possible studies.
All identified abstracts were carefully reviewed by two investigators (J. Q. Luo, H. Ren) independently for eligibility. The inclusion criteria were as follows: (i) case‐control design, regardless of sample size; (ii) study assessing the associations between Q192R (rs662) and L55M (rs854560) of PON1 gene and type 2 diabetes; (iii) numbers for the PON1 genotypes could be available or calculated in case and control groups; and (iv) genotype distribution in the controls was in Hardy‐Weinberg equilibrium (HWE). If the two investigators (J. Q. Luo, H. Ren) disagreed about the eligibility of an article, it was resolved by consensus with a third reviewer (M. Z. Liu).
Data extraction
For the eligible articles included in this study, data were also extracted by two reviewers (J. Q. Luo, H. Ren), who reached a consensus on all of the data extraction items. The following information was extracted from each study: name of the first author, publication year, country of the study, ethnicity of the population, genotype and allele distributions in case and control groups, and also sample size, mean age and gender distribution in case and control groups.
Statistical analysis
The goodness‐of‐fit chi‐square analysis was used to test the HWE of the genotype distribution of controls. The distribution was considered deviated significantly from HWE with P < 0.05. The pooled odds ratio (OR) with 95% confidence interval (CI) was used to evaluate the strength of association in the allelic, homozygous, heterozygous, recessive and dominant models, respectively. The statistical significance of the pooled estimates of the OR was determined by the Z test. The Cochran's Q test and I 2 metric were performed to examine the possibility of between‐study heterogeneity. Heterogeneity was considered to be statistically significant at P < 0.05 for the Q statistic and I 2 > 50% for the I 2 metric 13. If substantial heterogeneity existed, random effect model (the DerSimonian and Laird method) was selected as the pooling method. Otherwise, the fixed‐effect model (the Mantel‐Haenszel method) was adopted. Subgroup analysis based on ethnicity (categorized as Europeans, East Asians, South Asians and Canadian Aboriginal) and meta‐regression with restricted maximum likelihood estimation were conducted to assess the sources of heterogeneity across the studies. Potential publication bias was assessed by Begg's test and Egger's test 14, 15, with P < 0.05 considered representative of significant publication bias. All statistical analyses were performed with STATA version 12.0 (Stata, College Station, TX, USA).
Results
Description of eligible studies
The initial screening yielded 332 articles, and 1 article was found to be eligible by reviewing the cited references. A total of 111 articles were excluded because of duplicate publication. Then, 57 articles were excluded from screening based on the titles and/or abstracts. Finally, 37 articles 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52 involving 50 eligible studies were included in the current meta‐analysis according to the study inclusion criteria (Fig. 1). All the included articles were case‐control designs with sample sizes varied from 61 to 593. A total of 34 and 16 eligible studies were identified for the PON1 Q192R (rs662) and L55M (rs854560) polymorphism, respectively. The general characteristics of the studies included in the meta‐analysis are presented in Table 1.
Figure 1.
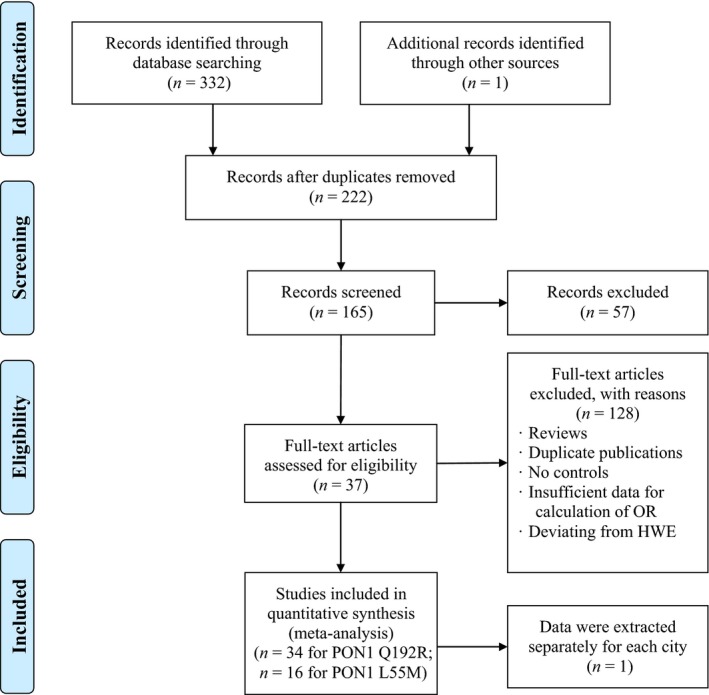
Flow diagram of the search strategy and study selection. The terms ‘n’ in the boxes represent the number of corresponding studies.
Table 1.
Characteristics of the included studies of the association of the PON1 Q192R and L55M genetic polymorphism with type 2 diabetes
| Study per SNP | Year | Country (Population)a | Male/Female | Age(years) | Sample sizeb | Case genotypes or allelesc | Control genotypes or allelesc | MAFd | HWE Pe | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case | Control | Case | Control | 11 | 12 | 22 | 1 | 2 | 11 | 12 | 22 | 1 | 2 | ||||||
| PON1 Q 192R | |||||||||||||||||||
| Mackness | 1998 | United Kingdom(EUR) | 162/90 | 147/135 | 59.1 ± 11.3 | 42.2 ± 12.2 | 252/282 | 117 | 99 | 34 | 333 | 167 | 156 | 99 | 24 | 411 | 147 | 0.263 | 0.153 |
| Sakai | 1998 | Japan(EAS) | 65/74 | 179/61 | 61.7 ± 13.6 | 48.0 ± 8.7 | 139/240 | 14 | 63 | 62 | 91 | 187 | 24 | 102 | 114 | 150 | 330 | 0.688 | 0.866 |
| Fanella | 2000 | Canada | NA | NA | 44.5 ± 15.4 | 25.4 ± 12.6 | 115/478 | 74 | 36 | 5 | 184 | 46 | 276 | 175 | 27 | 727 | 229 | 0.240 | 0.915 |
| Koch(a) | 2001 | Germany(EUR) | NA | NA | NA | NA | 39/202 | 25 | 12 | 2 | 62 | 16 | 102 | 87 | 13 | 291 | 113 | 0.280 | 0.328 |
| Koch(b) | 2001 | Germany(EUR) | NA | NA | NA | NA | 36/113 | 17 | 19 | 0 | 53 | 19 | 62 | 45 | 6 | 169 | 57 | 0.252 | 0.554 |
| Sampson | 2001 | United Kingdom(EUR) | 18/22 | 14/16 | 56.6 ± 7.3 | 52.0 ± 7.4 | 40/30 | 17 | 16 | 2 | 50 | 20 | 12 | 11 | 3 | 35 | 17 | 0.327 | 0.844 |
| Letellier | 2002 | France(EUR) | 57/39 | 52/53 | 56.8 ± 10.7 | 46.7 ± 10.9 | 167/105 | 92 | 67 | 8 | 251 | 83 | 40 | 50 | 14 | 130 | 78 | 0.375 | 0.794 |
| Hu YM | 2003 | China(EAS) | 95/57 | 83/45 | 58.5 ± 12.1 | 54 ± 11.5 | 152/128 | 30 | 77 | 45 | 137 | 167 | 25 | 74 | 29 | 124 | 132 | 0.516 | 0.075 |
| Zhang | 2003 | Japan(EAS) | 39/17 | 61/28 | 64.5 ± 7.5 | 62.7 ± 8.3 | 56/89 | 10 | 23 | 23 | 43 | 69 | 7 | 33 | 49 | 47 | 131 | 0.736 | 0.665 |
| Ma RX | 2003 | China(EAS) | 40/40 | 55/49 | 63.0 ± 8.0 | 64.0 ± 7.0 | 176/104 | 16 | 84 | 76 | 116 | 236 | 22 | 48 | 34 | 92 | 116 | 0.558 | 0.511 |
| Wang Y | 2003 | China(EAS) | 24/12 | 29/9 | 64.8 ± 11.9 | 70.8 ± 10.8 | 75/38 | 18 | 41 | 16 | 77 | 73 | 15 | 18 | 5 | 48 | 28 | 0.368 | 0.912 |
| Pu X | 2003 | China(EAS) | 14/16 | 55/45 | 67.0 ± 5.0 | 64.0 ± 4.0 | 100/100 | 6 | 64 | 30 | 76 | 124 | 18 | 52 | 30 | 88 | 112 | 0.56 | 0.581 |
| Hao YL | 2003 | China(EAS) | 50/55 | 44/36 | 54.5 ± 10.2 | 51.6 ± 6.3 | 187/80 | 26 | 86 | 75 | 138 | 236 | 6 | 32 | 42 | 44 | 116 | 0.725 | 0.978 |
| Ren T | 2003 | China(EAS) | 65/55 | 57/26 | NA | NA | 112/83 | 22 | 95 | 78 | 139 | 251 | 9 | 42 | 29 | 60 | 100 | 0.625 | 0.283 |
| Li SY | 2004 | China(EAS) | 22/14 | 21/12 | 56.0 ± 8.0 | 57.0 ± 11.0 | 63/33 | 16 | 24 | 23 | 56 | 70 | 13 | 13 | 7 | 39 | 27 | 0.409 | 0.287 |
| Zhang Z | 2004 | China(EAS) | 30/26 | 49/31 | 63.6 ± 11.4 | 63.7 ± 11.5 | 116/80 | 16 | 41 | 59 | 73 | 159 | 12 | 41 | 27 | 65 | 95 | 0.594 | 0.577 |
| Deng YG | 2004 | China(EAS) | 37/43 | 45/45 | 60.1 ± 2.7 | 54.8 ± 3.7 | 169/90 | 20 | 57 | 90 | 97 | 237 | 13 | 44 | 33 | 70 | 110 | 0.611 | 0.786 |
| Sun YD | 2005 | China(EAS) | 92/85 | 50/47 | 64.5 ± 10.3 | 62.4 ± 10.9 | 162/97 | 53 | 161 | 95 | 267 | 351 | 14 | 56 | 27 | 84 | 110 | 0.567 | 0.083 |
| Mastorikou | 2006 | United Kingdom(EUR) | 21/15 | 10/9 | 57.7 ± 5.2 | 57.7 ± 4.8 | 36/19 | NA | NA | NA | 48 | 24 | NA | NA | NA | 28 | 10 | 0.263 | NA |
| Qi L | 2007 | China(EAS) | 44/49 | 35/54 | 56.6 ± 7.0 | 57.9 ± 6.8 | 183/89 | 32 | 97 | 54 | 161 | 205 | 18 | 42 | 29 | 78 | 100 | 0.562 | 0.695 |
| Shi GH | 2007 | China(EAS) | 49/43 | 43/38 | 60.9 ± 7.3 | 62.4 ± 6.3 | 179/81 | 33 | 69 | 77 | 135 | 223 | 12 | 38 | 31 | 62 | 100 | 0.617 | 0.949 |
| Irace | 2008 | Italy(EUR) | NA | NA | 55.2 ± 9.2 | 55.9 ± 6.6 | 118/65 | 64 | 42 | 12 | 170 | 66 | 26 | 31 | 8 | 83 | 47 | 0.362 | 0.790 |
| Unür | 2008 | Turkey(EUR) | 20/31 | 27/26 | 52.5 ± 5.6 | 55.5 ± 8.0 | 51/53 | 31 | 14 | 6 | 76 | 26 | 25 | 22 | 6 | 72 | 34 | 0.321 | 0.730 |
| Flekac | 2008 | Czech(EUR) | 114/132 | 55/45 | 58 ± 18 | 41 ± 9 | 246/110 | 177 | 64 | 5 | 418 | 74 | 32 | 54 | 24 | 118 | 102 | 0.464 | 0.892 |
| Gorshunska | 2009 | Ukraine(EUR) | 28/68 | 86/47 | 56.2 ± 1.4 | 55.3 ± 2.5 | 96/123 | 55 | 25 | 16 | 135 | 57 | 48 | 64 | 11 | 160 | 86 | 0.350 | 0.110 |
| Ergun | 2011 | Turkey(EUR) | NA | NA | 59 ± 9.63 | 47 ± 6.53 | 171/80 | 91 | 50 | 30 | 232 | 110 | 38 | 31 | 11 | 107 | 53 | 0.331 | 0.262 |
| Bhaskar | 2011 | India(SAS) | NA | NA | NA | NA | 310/120 | 71 | 184 | 55 | 326 | 294 | 40 | 66 | 14 | 146 | 94 | 0.392 | 0.091 |
| Gupta | 2011 | India(SAS) | 126/124 | 151/149 | 47.4 ± 11.3 | 43.1 ± 10.7 | 250/300 | 81 | 126 | 43 | 288 | 212 | 168 | 108 | 24 | 444 | 156 | 0.26 | 0.264 |
| Chen XJ | 2011 | China(EAS) | 50/47 | 55/50 | 59.9 ± 10.6 | 58.7 ± 5.7 | 210/105 | 23 | 109 | 78 | 155 | 265 | 12 | 58 | 35 | 82 | 128 | 0.610 | 0.100 |
| Elnoamany | 2012 | Egypt(EUR) | 33/12 | 30/10 | 51.11 ± 6.7 | 50.19 ± 5.5 | 93/40 | 42 | 28 | 23 | 112 | 74 | 25 | 13 | 2 | 63 | 17 | 0.213 | 0.855 |
| Zheng YQ | 2012 | China(EAS) | 51/39 | 70/66 | 57.1 ± 12.0 | 45.5 ± 13.3 | 184/136 | 21 | 66 | 97 | 108 | 260 | 19 | 57 | 60 | 95 | 177 | 0.651 | 0.363 |
| Gokcen | 2013 | Turkey(EUR) | 20/30 | 16/14 | 60.8 ± 9.4 | 54.2 ± 8.1 | 50/30 | 18 | 25 | 7 | 61 | 39 | 8 | 14 | 8 | 30 | 30 | 0.500 | 0.715 |
| Shao ZY | 2014 | China(EAS) | 94/83 | 111/95 | 63.3 ± 10.9 | 61.3 ± 11.0 | 379/206 | 50 | 173 | 156 | 273 | 485 | 35 | 94 | 77 | 164 | 248 | 0.602 | 0.493 |
| Du WL | 2015 | China(EAS) | 28/33 | 28/36 | 54.9 ± 12.7 | 37.7 ± 14.5 | 125/64 | 7 | 43 | 75 | 57 | 193 | 5 | 22 | 37 | 32 | 96 | 0.75 | 0.505 |
| PON1 L55M | |||||||||||||||||||
| Ikeda | 1998 | Japan(EAS) | 53/55 | 82/79 | 58 ± 7 | 57 ± 8 | 108/161 | 95 | 10 | 3 | 200 | 16 | 142 | 19 | 0 | 303 | 19 | 0.059 | 0.426 |
| Malin | 1999 | Finnish(EUR) | NA | NA | NA | NA | 93/106 | 33 | 49 | 11 | 115 | 71 | 38 | 54 | 14 | 130 | 82 | 0.387 | 0.447 |
| Fanella | 2000 | Canada | NA | NA | 44.5 ± 15.4 | 25.4 ± 12.6 | 115/478 | 113 | 2 | 0 | 228 | 2 | 471 | 7 | 0 | 949 | 7 | 0.007 | 0.872 |
| Letellier | 2002 | France(EUR) | 57/39 | 52/53 | 56.8 ± 10.7 | 46.7 ± 10.9 | 167/105 | 65 | 81 | 20 | 211 | 121 | 40 | 52 | 12 | 132 | 76 | 0.366 | 0.426 |
| Ren T | 2003 | China(EAS) | 65/55 | 57/26 | NA | NA | 184/83 | 177 | 7 | 0 | 361 | 7 | 70 | 2 | 0 | 142 | 2 | 0.013 | 0.905 |
| Agachan | 2004 | Turkish(EUR) | 122/91 | 57/59 | 59.9 ± 11.6 | 58.6 ± 16.0 | 213/116 | 111 | 86 | 8 | 308 | 102 | 51 | 51 | 7 | 153 | 65 | 0.298 | 0.218 |
| Sampson | 2005 | United Kingdom(EUR) | 38/20 | 18/32 | NA | NA | 58/50 | 26 | 32 | 32 | NA | NA | 21 | 28 | 28 | NA | NA | NA | NA |
| Mastorikou | 2006 | United Kingdom(EUR) | 21/15 | 10/9 | 57.7 ± 5.2 | 57.7 ± 4.8 | 36/19 | NA | NA | NA | 49 | 23 | NA | NA | NA | 24 | 14 | 0.368 | NA |
| Sun YD | 2006 | China(EAS) | 92/85 | 50/47 | 64.5 ± 10.3 | 62.4 ± 10.9 | 294/91 | 121 | 150 | 23 | 392 | 196 | 38 | 45 | 8 | 121 | 61 | 0.335 | 0.296 |
| Shao HQ | 2006 | China(EAS) | 29/21 | 60/60 | 61.5 ± 3.3 | 55.0 ± 2.3 | 92/120 | 85 | 7 | 0 | 177 | 7 | 109 | 11 | 0 | 229 | 11 | 0.046 | 0.599 |
| Flekac | 2008 | Czech(EUR) | 114/132 | 55/45 | 58 ± 18 | 41 ± 9 | 246/110 | 84 | 118 | 44 | 286 | 206 | 30 | 55 | 25 | 115 | 105 | 0.477 | 0.983 |
| Unür | 2008 | Turkey(EUR) | 20/31 | 27/26 | 52.5 ± 5.6 | 55.5 ± 8.0 | 51/53 | 43 | 5 | 3 | 91 | 11 | 28 | 20 | 5 | 76 | 30 | 0.283 | 0.609 |
| Altuner | 2011 | Turkey(EUR) | 56/44 | 21/29 | 54.4 ± 2.8 | 44.04 ± 1.1 | 100/50 | 43 | 44 | 13 | 130 | 70 | 21 | 25 | 4 | 67 | 33 | 0.33 | 0.355 |
| Ergun | 2011 | Turkish(EUR) | NA | NA | 59 ± 9.6 | 47 ± 6.5 | 171/80 | 35 | 45 | 91 | 115 | 227 | 16 | 30 | 34 | 62 | 98 | 0.613 | 0.060 |
| Gupta | 2011 | India(SAS) | 126/124 | 151/149 | 47.4 ± 11.3 | 43.1 ± 10.7 | 250/300 | 176 | 69 | 5 | 421 | 79 | 193 | 101 | 6 | 487 | 113 | 0.188 | 0.080 |
| Zheng YQ | 2012 | China(EAS) | 51/39 | 70/66 | 57.1 ± 12.0 | 45.5 ± 13.3 | 184/136 | 168 | 15 | 1 | 351 | 17 | 124 | 11 | 1 | 259 | 13 | 0.048 | 0.194 |
| Shao ZY | 2014 | China(EAS) | 94/83 | 111/95 | 63.3 ± 10.9 | 61.3 ± 11.0 | 379/206 | 339 | 34 | 6 | 712 | 46 | 184 | 20 | 2 | 388 | 24 | 0.058 | 0.099 |
The population codes of EAS, EUR and SAS mean East Asian, European and South Asian, respectively.
Sample size means the case‐control groups.
For the PON1 Q192R, 11: QQ, 12: QR, 22: RR; for the PON1 L55M, 11: LL, 12:LM, 22: MM.
MAF, minor allele frequency; NA, not available.
HWE, Hardy‐Weinberg equilibrium.
Quantitative synthesis of the association between PON1 Q912R polymorphism and T2DM
The results of the meta‐analysis of PON1 Q912R polymorphism are summarized in detail in Table 2 and Figure 2. In the overall population, the pooled meta‐analysis revealed that there were no significant associations between the PON1 Q912R genetic polymorphism and T2DM under all genetic models: allelic (OR = 1.02, 95% CI = 0.87–1.20; P = 0.786), homozygous (OR = 1.08, 95% CI = 0.81–1.45; P = 0.596), heterozygous (OR = 0.93, 95% CI = 0.75–1.17; P = 0.544), recessive (OR = 1.12, 95% CI = 0.92–1.35; P = 0.259) and dominant (OR = 0.99, 95% CI = 0.78–1.26; P = 0.921).
Table 2.
Summary of meta‐analysis of the association of the PON1 Q192R and L55M genetic polymorphism with type 2 diabetes
| Genetic modela | PON1 R192R | PON1 L55M | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pooled OR (95%CI) | Z | P b | N c | Modeld | I 2% | P hetero | Pooled OR(95%CI) | Z | P b | N c | Modeld | I 2% | P hetero | |
| Allelic | 1.02 (0.87–1.20) | 0.27 | 0.786 | 34 | R | 81.8% | 0.000 | 0.91 (0.81–1.02) | 1.56 | 0.118 | 16 | F | 2.3% | 0.427 |
| EUR subgroup | 0.80 (0.56–1.16) | 1.16 | 0.246 | 13 | R | 87.1% | 0.000 | 0.89 (0.77–1.03) | 1.52 | 0.129 | 8 | F | 45.7% | 0.075 |
| Canadian Aboriginal | 0.79 (0.56–1.13) | 1.27 | 0.203 | 1 | NA | NA | NA | 1.19 (0.25–5.76) | 0.22 | 0.830 | 1 | NA | NA | NA |
| SAS subgroup | 1.73 (1.17–2.56) | 2.72 | 0.007b | 2 | R | 74.8% | 0.046 | 0.81 (0.59–1.11) | 1.32 | 0.187 | 1 | NA | NA | NA |
| EAS subgroup | 1.14 (1.01–1.28) | 2.1 | 0.036b | 18 | R | 43.4% | 0.026 | 1.03 (0.81–1.31) | 0.22 | 0.825 | 6 | F | 0.0% | 0.978 |
| Recessive | 1.12 (0.92–1.35) | 1.13 | 0.259 | 33 | R | 62.6% | 0.000 | 1.05 (0.81–1.35) | 0.35 | 0.729 | 12 | F | 0.0% | 0.630 |
| EUR subgroup | 0.77 (0.41–1.46) | 0.80 | 0.425 | 12 | R | 77.2% | 0.000 | 1.02 (0.76–1.35) | 0.11 | 0.912 | 7 | F | 0.0% | 0.427 |
| Canadian Aboriginal | 0.76 (0.29–2.02) | 0.55 | 0.580 | 1 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| SAS subgroup | 2.03 (1.35–3.05) | 3.39 | 0.001b | 2 | F | 0.0% | 0.365 | 1.00 (0.30–3.32) | 0.00 | 1.000 | 1 | NA | NA | NA |
| EAS subgroup | 1.18 (1.04–1.33) | 2.6 | 0.009b | 18 | F | 33.4% | 0.084 | 1.25 (0.63–2.47) | 0.63 | 0.529 | 4 | F | 0.0% | 0.405 |
| Dominant | 0.99 (0.78–1.26) | 0.10 | 0.921 | 33 | R | 78.4% | 0.000 | 0.85 (0.73–0.99) | 2.15 | 0.032b | 16 | F | 0.0% | 0.628 |
| EUR subgroup | 0.69 (0.45–1.06) | 1.70 | 0.089 | 12 | R | 83.5% | 0.000 | 0.80 (0.65–0.99) | 2.10 | 0.036b | 8 | F | 31.9% | 0.173 |
| Canadian Aboriginal | 0.76 (0.50–1.16) | 1.29 | 0.197 | 1 | NA | NA | NA | 1.19 (0.24–5.81) | 0.22 | 0.829 | 1 | NA | NA | NA |
| SAS subgroup | 2.26 (1.72–2.98) | 5.78 | 0.000b | 2 | F | 57.9% | 0.123 | 0.76 (0.53–1.09) | 1.51 | 0.132 | 1 | NA | NA | NA |
| EAS subgroup | 1.18 (0.99–1.39) | 1.88 | 0.060 | 18 | F | 30.4% | 0.108 | 1.00 (0.75–1.33) | 0 | 0.997 | 6 | F | 0.0% | 0.997 |
| Homozygous | 1.08 (0.81–1.45) | 0.53 | 0.596 | 33 | R | 72.1% | 0.000 | 0.92 (0.69–1.23) | 0.56 | 0.577 | 12 | F | 0.0% | 0.700 |
| EUR subgroup | 0.63 (0.30–1.32) | 1.22 | 0.222 | 12 | R | 81.6% | 0.000 | 0.85 (0.61–1.19) | 0.94 | 0.348 | 7 | F | 0.0% | 0.562 |
| Canadian Aboriginal | 0.69 (0.26–1.86) | 0.73 | 0.463 | 1 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| SAS subgroup | 3.01 (1.93–4.67) | 4.88 | 0.000b | 2 | F | 21.2% | 0.260 | 0.91 (0.27–3.05) | 0.15 | 0.883 | 1 | NA | NA | NA |
| EAS subgroup | 1.28 (1.06–1.54) | 2.54 | 0.011b | 18 | F | 34.5% | 0.075 | 1.28 (0.64–2.59) | 0.7 | 0.487 | 4 | F | 0.0% | 0.431 |
| Heterozygous | 0.93 (0.75–1.17) | 0.61 | 0.544 | 33 | R | 71.4% | 0.000 | 0.82 (0.70–0.97) | 2.39 | 0.017b | 15 | F | 0.0% | 0.609 |
| EUR subgroup | 0.66 (0.45–0.98) | 2.09 | 0.037b | 12 | R | 75.8% | 0.000 | 0.77 (0.61–0.97) | 2.24 | 0.025b | 7 | F | 37.7% | 0.141 |
| Canadian Aboriginal | 0.77 (0.49–1.19) | 1.18 | 0.239 | 1 | NA | NA | NA | 1.19 (0.24–5.81) | 0.22 | 0.829 | 1 | NA | NA | NA |
| SAS subgroup | 2.07 (1.54–2.76) | 4.89 | 0.000b | 2 | F | 49.1% | 0.161 | 0.75 (0.52–1.08) | 1.54 | 0.124 | 1 | NA | NA | NA |
| EAS subgroup | 1.09 (0.91–1.30) | 0.95 | 0.341 | 18 | F | 18.9% | 0.228 | 0.96 (0.72–1.29) | 0.28 | 0.778 | 6 | F | 0.0% | 0.984 |
The population codes of EAS, EUR and SAS mean East Asian, European and South Asian, respectively.
P < 0.05.
N means the number of eligible studies for the meta‐analysis.
F, fixed‐effects model; NA, not available; R, random‐effects model.
Figure 2.
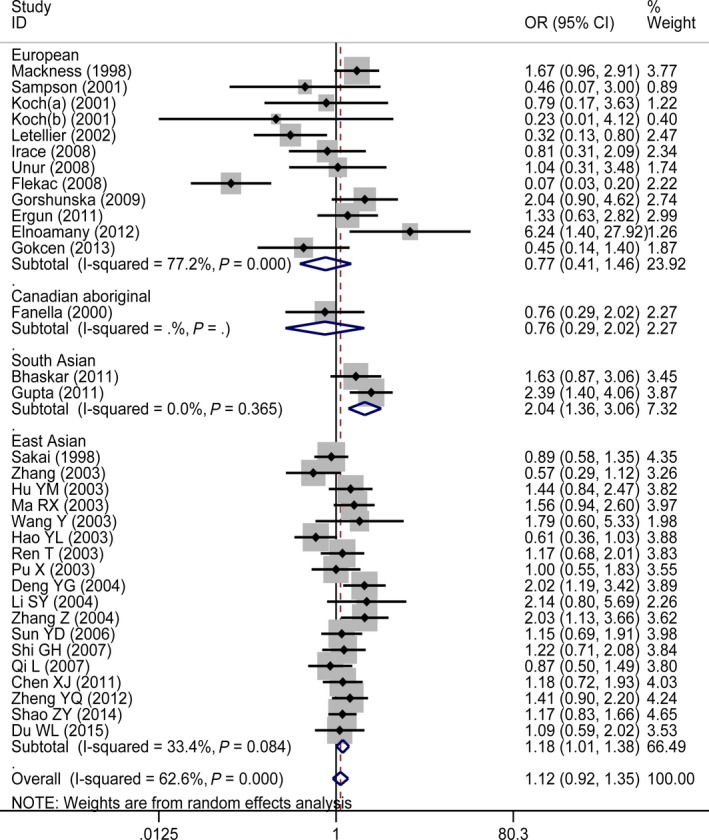
Forest plot for PON1 Q192R polymorphism under a recessive genetic model stratified by ethnicity in studies with type 2 diabetes patients.
When we performed subgroup analyses stratified by ethnicity, the distinct effects in different ethnic populations were observed under all genetic models. Significant associations between PON1 Q912R genetic polymorphism and T2DM presented in the South Asian subgroup (under all genetic models) and East Asian subgroup (under four genetic models), while no significant associations were shown in the Canadian Aboriginal subgroup and in the European subgroup under the allelic, homozygous, recessive and dominant genetic models. By contrast, the significant association for the European subgroup under the heterozygous genetic model showed the 192R allele represented a protective factor of T2DM (OR = 0.66, 95% CI = 0.45–0.98; P = 0.037), but a risk factor for T2DM in South Asian subgroup.
Quantitative synthesis of the association between PON1 L55M polymorphism and T2DM
The results of the meta‐analysis of PON1 L55M polymorphism are summarized in detail in Table 2 and Figure 3. In the overall population, the associations between the PON1 L55M genetic polymorphism and T2DM did not reach statistically significant under the allelic genetic model (OR = 0.91, 95% CI = 0.81–1.02; P = 0.118), homozygous genetic model (OR = 0.92, 95% CI = 0.69–1.23; P = 0.577) and recessive genetic model (OR = 1.05, 95% CI = 0.81–1.35; P = 0.729). However, significant associations were found under a heterozygous genetic model (OR = 0.82, 95% CI = 0.70–0.97; P = 0.017) and a dominant genetic model (OR = 0.85, 95% CI = 0.73–0.99; P = 0.032).
Figure 3.
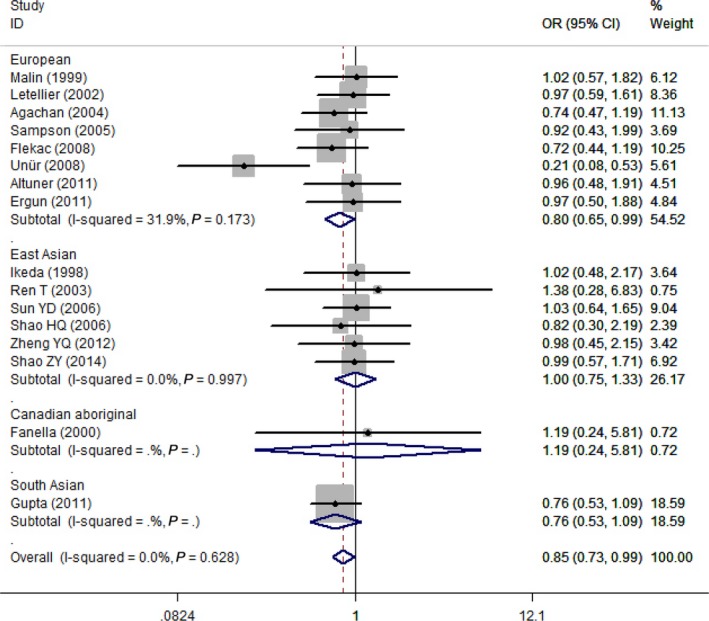
Forest plot for PON1 L55M polymorphism under a dominant genetic model stratified by ethnicity in studies with type 2 diabetes patients.
In subgroup analyses based on ethnicity, the distinct effects in different ethnic populations were also presented for the PON1 L55M genetic polymorphism. There were significant protective effects of L allele on T2DM in the European subgroup under the heterozygous (OR = 0.77, 95% CI = 0.61–0.97; P = 0.025) and dominant (OR = 0.80, 95% CI = 0.65–0.99; P = 0.036) genetic models, while no significant results were found in the South Asian, East Asian and Canadian Aboriginal subgroup under all genetic models.
Sources of heterogeneity
There was significant heterogeneity in the overall meta‐analysis of PON1 Q912R polymorphism under all of the genetic models (P heterogeneity < 0.05, I 2 > 50%). Subgroup analysis stratified by ethnicity indicated that heterogeneity was significantly reduced in the South Asian and East Asian subgroup, while was increased in the European subgroup. Therefore, ethnicity may be one of the sources of heterogeneity between studies for the PON1 Q912R polymorphism in the overall meta‐analysis.
Because substantial heterogeneity still existed in the European subgroup under all genetic models, meta‐regression was used to explore the source of this heterogeneity. The following three covariates were taken into consideration: publication year, MAF (minor allele frequency) in controls and sample size in the subsequent meta‐regression (Table 3). The results of meta‐regression analysis showed that MAF in the control group could explain the observed between‐study heterogeneity. The proportion of between‐study variance explained by the MAF covariate ranges from 67.81 to 93.06%, depending on the genetic models. However, no significant effects were accounted for by the covariates sample size and publication year under all genetic models.
Table 3.
The meta‐regression results among the European population under all genetic model for the PON1 Q192R genetic polymorphism
| Genetic model | Covariates | Coefficient | Standard Error | T‐value | P‐value | 95% confidence interval | Adjusted R‐squared |
|---|---|---|---|---|---|---|---|
| Heterozygous | MAF in controls | −6.03742 | 1.641809 | −3.68 | 0.004a | −9.6956∼−2.37924 | 81.00% |
| Sample size | −0.00028 | 0.001466 | −0.19 | 0.854 | −0.00354∼0.002991 | −14.64% | |
| Publication year | −0.04524 | 0.03562 | −1.27 | 0.233 | −0.12461∼0.034129 | 13.01% | |
| Allelic | MAF in controls | −6.41229 | 1.281261 | −5 | 0.000a | −9.23233∼−3.59225 | 80.80% |
| Sample size | −0.00045 | 0.001437 | −0.32 | 0.759 | −0.00366∼0.002749 | −11.10% | |
| Publication year | −0.00646 | 0.037309 | −0.17 | 0.866 | −0.08858∼0.075655 | −9.84% | |
| Homozygous | MAF in controls | −12.2995 | 3.278749 | −3.75 | 0.004a | −19.605∼−4.99395 | 73.94% |
| Sample size | −0.00058 | 0.003051 | −0.19 | 0.853 | −0.00738∼0.006216 | −13.55% | |
| Publication year | 0.023242 | 0.081639 | 0.28 | 0.782 | −0.15866∼0.205145 | −13.07% | |
| Dominant | MAF in controls | −7.93603 | 1.488279 | −5.33 | 0.000a | −11.2521∼−4.61994 | 93.06% |
| Sample size | −0.00049 | 0.001638 | −0.3 | 0.771 | −0.00414∼0.003161 | −12.29% | |
| Publication year | −0.02767 | 0.042206 | −0.66 | 0.527 | −0.12171∼0.066367 | −4.49% | |
| Recessive | MAF in controls | −10.0113 | 3.071858 | −3.26 | 0.009a | −16.8558∼−3.16679 | 67.81% |
| Sample size | −0.00047 | 0.002671 | −0.18 | 0.864 | −0.00642∼0.005483 | −14.87% | |
| Publication year | 0.035213 | 0.071726 | 0.49 | 0.634 | −0.1246∼0.19503 | −13.82% |
P < 0.05.
MAF, minor allele frequency; Coefficient: regression coefficient. The regression coefficients were the estimated increase in the lnOR per unit increase in the covariates.
In contrast, no significant heterogeneity in the overall meta‐analysis of PON1 L55M polymorphism was showed under all genetic models (P heterogeneity > 0.1, I 2 = 0%). Subgroup analysis stratified by ethnicity also indicated that no substantial between‐study heterogeneity was found in the Asian subgroup (P heterogeneity > 0.1, I 2 = 0%) and in the European subgroup (P heterogeneity > 0.05, I 2 < 50%) under all genetic models.
Publication bias evaluation
Publication bias of the individual articles was evaluated by using the Begg's funnel plot (Fig. 4) and Egger's test. For the PON1 Q192R meta‐analysis (Fig. 4A), no obvious publication bias was visualized in the shape of the funnel plot under all genetic models. Additionally, no evidence of significant publication bias was detected by the Egger's test (P = 0.257 for allelic genetic model; P = 0.452 for heterozygous genetic model; P = 0.527 for dominant genetic model; and P = 0.197 for recessive genetic model). However, there was marginal significant publication bias for the homozygous genetic model (P = 0.047).
Figure 4.
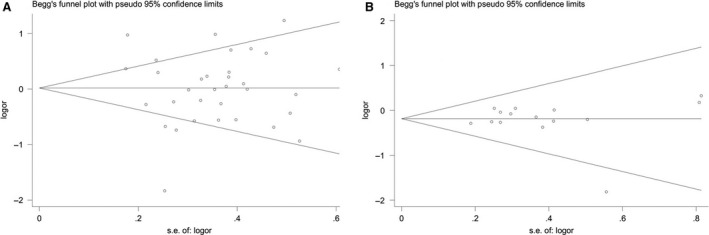
Begg's funnel plot for studies of the association between type 2 diabetes and PON1 Q192R polymorphism under a dominant genetic model (A) and PON1 L55M polymorphism under a heterozygous genetic model (B).
For the PON1 L55R meta‐analysis (Fig. 4B), there is also no obvious publication bias in the shape of the funnel plot under all genetic models. No evidence of significant publication bias was also detected by the Egger's test (P = 0.961 for allelic genetic model; P = 0.719 for heterozygous genetic model; P = 0.309 for homozygous genetic model; P = 0.871 for dominant genetic model; and P = 0.628 for recessive genetic model) yet.
Discussion
So far, the associations between PON1 genetic polymorphisms and T2DM were conflicting in the previous studies. This is partly because some previous case‐control studies have been too small to be reliable. Thus, our meta‐analysis could overcome the limitations of single study by pooling the individual dataset and provide more reliable results.
In the overall meta‐analysis of the PON1 Q192R polymorphism, no significant association, but strong between‐study heterogeneity, was observed. To address the substantial heterogeneity, we divided the total samples into four subgroups, that is white European, Canadian Aboriginal, South and East Asians. Stratified analyses by ethnicity yielded a significant association of the PON1 Q192R polymorphism with T2DM in South Asian and East Asian populations and, conversely, no association of the PON1 Q192R polymorphism with T2DM in European populations under the allelic, homozygous, recessive and dominant genetic models. In addition, the 192R allele was a susceptible factor of T2DM in the Asian population but represented a protective factor of T2DM in European population under a heterozygous genetic model.
In the overall meta‐analysis of the PON1 L55M polymorphism, no significance between‐study heterogeneity was observed. The distinct effects across different ethnic backgrounds also presented in the subgroup analysis based on ethnicity. For example, significant protective effects of the 55M allele on T2DM under the heterozygous and dominant genetic models were found in the European population, while no significant results in the Asian populations under all genetic models. Interestingly, the associations of the two PON1 SNPs in our study were generally very similar in South and East Asians, although Asia is known to harbour genetically different origins 53. In Canadian population, only one study investigated the association between the two PON1 SNPs and risk of T2DM, and no significant associations were found in all genetic models. Therefore, it was inferred that the 192R or 55M allele may decrease the risk of developing T2DM in European ancestry population, whereas the 192R increase the risk of T2DM in the South Asian and East Asian populations.
To our knowledge, this is the largest study to underline the importance of ethnicity in the association between PON1 genetic variations and T2DM by a comprehensive meta‐analysis. The question remaining to be addressed is how the PON1 Q192R and L55M variants can exert an impact on T2DM with ethnic difference. One potential explanation is that different populations might have experienced very diverse lifestyle and environmental factors during their long‐period evolution. The PON1 activity may be influenced by several environmental and lifestyle impacts, such as cigarette smoking 54, alcohol intake 55 and physical activity 56. Another possible explanation may be the ethical differences in the distribution of the PON1 Q192R and L55M (rs854560) polymorphisms. Nevertheless, the precise mechanism deserves to be investigated in the future.
According to the included studies among different countries 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52 and the 1000 genomes database (https://www.ncbi.nlm.nih.gov/variation/tools/1000genomes/), there are huge racial and regional differences in the distribution of the PON1 Q192R (rs662) and L55M (rs854560) genetic polymorphisms (Fig. 5). For the PON1 Q192R polymorphism, the R allele predominates in the East Asian populations (>60%), which is significantly higher than in the South Asian populations (about 40%) and the European populations (<35%). For the PON1 L55M polymorphism, the M allele frequency is rare in the East Asian populations (<5%), which is significantly lower than in the South Asian populations (about 20%) and European populations (>30%). Such heterogeneous genetic backgrounds could be, at least in part, responsible for the heterogeneity of effect on the risk of T2DM detected in our overall population meta‐analysis. Furthermore, subgroup analysis stratified by ethnicity also indicated that the heterogeneity in the Asian group was significantly decreased.
Figure 5.
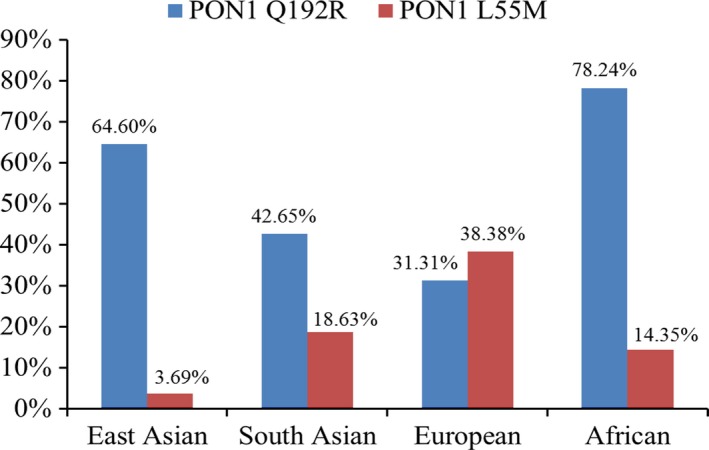
The frequency of PON1 Q192R and L55M among the different ethnicities. The data were summarized according to the 1000 genomes database. East Asian referred to the Chinese and Japanese; South Asian was from India; European referred to Utah Residents (CEPH) with Northern and Western European Ancestry; African was from Yoruba in Ibadan, Nigeria.
The meta‐analysis results in the current study should be interpreted with particular caution when large between‐study heterogeneity existed. Obvious heterogeneity was present in all the genetic models for the PON1 Q192R polymorphism in the European population subgroup. Meta‐regression was performed to evaluate the potentially important covariates exerting substantial impact on heterogeneity. Our findings have proved that the proportion of heterogeneity explained by the MAF in controls can reach as high as 93.06%. One of the reasons may be the small number of subjects in the control group. For example, the study of Elnoamany et al. 45 included 40 control subjects and the MAF of PON1 Q192R was 0.213, while the study of Gokcen et al. 47. included 30 control subjects and the MAF of PON1 Q192R was 0.5. Accordingly, studies with large sample size are needed to be investigated in the future.
There are some shortcomings in our current meta‐analysis. First, our included studies were limited to English and Chinese language, with some data published in other languages excluded, which may lead to some publication bias and thus affect the pooled results in the meta‐analysis. Second, although there are 34 eligible studies for the PON1 Q192R polymorphism meta‐analysis and 16 eligible studies for the PON1 L55M polymorphism, the populations were restricted to Asians, Europeans and Canadian Aboriginals. Studies from other populations should be conducted to confirm the findings. Last but not the least, the information about exposure to environmental substrates was not available in the included studies. This may explain some between‐study heterogeneity in our meta‐analysis. In addition, the gene×environment interactions are needed to be further evaluated in the future.
In conclusion, we have firmly established that the PON1 genetic polymorphisms (Q192R and L55M) play important roles in the risk of T2DM with distinct effects across European and Asian populations. Further studies from other populations are needed to confirm these results.
Conflict of interest
The authors confirm that there are no conflicts of interest.
Acknowledgement
Conceived and designed the study: JQL and HR. Performed the search: JQL, HR and MZL. Analysed the data: JQL and HR. Contributed reagents/material/analysis tools: JQL, HR, MZL, PFF and DXX. Wrote the manuscript, reference collection, data management, statistical analyses, paper writing and study design: JQL.
Funding source: This work was supported by the National Natural Science Foundation of China (No. 81703623).
References
- 1. Collaboration NCDRF . Worldwide trends in diabetes since 1980: a pooled analysis of 751 population‐based studies with 4.4 million participants. Lancet. 2016; 387: 1513–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. GBD Disease and Injury Incidence and Prevalence Collaborators . Global, regional, and national incidence, prevalence, and years lived with disability for 310 diseases and injuries, 1990‐2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet. 2016; 388: 1545–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yang W, Lu J, Weng J, et al Prevalence of diabetes among men and women in China. N Engl J Med. 2010; 362: 1090–101. [DOI] [PubMed] [Google Scholar]
- 4. Qiu CJ, Ye XZ, Yu XJ, et al Association between FABP2 Ala54Thr polymorphisms and type 2 diabetes mellitus risk: a HuGE Review and Meta‐Analysis. J Cell Mol Med. 2014; 18: 2530–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Li YY, Wang XM, Lu XZ. KCNQ1 rs2237892 C→T gene polymorphism and type 2 diabetes mellitus in the Asian population: a meta‐analysis of 15,736 patients. J Cell Mol Med. 2014; 18: 274–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dullaart RP, de Vries R, Sluiter WJ, et al High plasma C‐reactive protein (CRP) is related to low paraoxonase‐I (PON‐I) activity independently of high leptin and low adiponectin in type 2 diabetes mellitus. Clin Endocrinol. 2009; 70: 221–6. [DOI] [PubMed] [Google Scholar]
- 7. Dullaart RP, Otvos JD, James RW. Serum paraoxonase‐1 activity is more closely related to HDL particle concentration and large HDL particles than to HDL cholesterol in Type 2 diabetic and non‐diabetic subjects. Clin Biochem. 2014; 47: 1022–7. [DOI] [PubMed] [Google Scholar]
- 8. Bhattacharyya T, Nicholls SJ, Topol EJ, et al Relationship of paraoxonase 1 (PON1) gene polymorphisms and functional activity with systemic oxidative stress and cardiovascular risk. JAMA. 2008; 299: 1265–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mackness B, Durrington PN, Mackness MI. Human serum paraoxonase. Gen Pharmacol. 1998; 31: 329–36. [DOI] [PubMed] [Google Scholar]
- 10. Mackness MI, Arrol S, Mackness B, et al Alloenzymes of paraoxonase and effectiveness of high‐density lipoproteins in protecting low‐density lipoprotein against lipid peroxidation. Lancet. 1997; 349: 851–2. [DOI] [PubMed] [Google Scholar]
- 11. Brophy VH, Jampsa RL, Clendenning JB, et al Effects of 5′ regulatory‐region polymorphisms on paraoxonase‐gene (PON1) expression. Am J Hum Genet. 2001; 68: 1428–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ikeda Y, Suehiro T, Inoue M, et al Serum paraoxonase activity and its relationship to diabetic complications in patients with non‐insulin‐dependent diabetes mellitus. Metabolism. 1998; 47: 598–602. [DOI] [PubMed] [Google Scholar]
- 13. Higgins JP, Thompson SG, Deeks JJ, et al Measuring inconsistency in meta‐analyses. BMJ. 2003; 327: 557–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Egger M, Davey Smith G, Schneider M, et al Bias in meta‐analysis detected by a simple, graphical test. BMJ. 1997; 315: 629–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994; 50: 1088–101. [PubMed] [Google Scholar]
- 16. Mackness B, Mackness MI, Arrol S, et al Serum paraoxonase (PON1) 55 and 192 polymorphism and paraoxonase activity and concentration in non‐insulin dependent diabetes mellitus. Atherosclerosis. 1998; 139: 341–9. [DOI] [PubMed] [Google Scholar]
- 17. Sakai T, Matsuura B, Onji M. Serum paraoxonase activity and genotype distribution in Japanese patients with diabetes mellitus. Intern Med. 1998; 37: 581–4. [DOI] [PubMed] [Google Scholar]
- 18. Malin R, Rantalaiho V, Huang XH, et al Association between M/L55‐polymorphism of paraoxonase enzyme and oxidative DNA damage in patients with type 2 diabetes mellitus and in control subjects. Hum Genet. 1999; 105: 179–80. [DOI] [PubMed] [Google Scholar]
- 19. Fanella S, Harris SB, Young TK, et al Association between PON1 L/M55 polymorphism and plasma lipoproteins in two Canadian aboriginal populations. Clin Chem Lab Med. 2000; 38: 413–20. [DOI] [PubMed] [Google Scholar]
- 20. Koch M, Hering S, Barth C, et al Paraoxonase 1 192 Gln/Arg gene polymorphism and cerebrovascular disease: Interaction with type 2 diabetes. Exp Clin Endocrinol Diabetes. 2001; 109: 141–5. [DOI] [PubMed] [Google Scholar]
- 21. Sampson MJ, Astley S, Richardson T, et al Increased DNA oxidative susceptibility without increased plasma LDL oxidizability in Type II diabetes: effects of alpha‐tocopherol supplementation. Clin Sci. 2001; 101: 235–41. [PubMed] [Google Scholar]
- 22. Letellier C, Durou MR, Jouanolle AM, et al Serum paraoxonase activity and paraoxonase gene polymorphism in type 2 diabetic patients with or without vascular complications. Diabetes Metab. 2002; 28: 297–304. [PubMed] [Google Scholar]
- 23. Hu Y, Tian H, Liu R. Gln‐Arg192 polymorphism of paraoxonase 1 is associated with carotid intima‐media thickness in patients of type 2 diabetes mellitus of Chinese. Diabetes Res Clin Pract. 2003; 61: 21–7. [DOI] [PubMed] [Google Scholar]
- 24. Zhang B, Eto S, Fan P, et al Paraoxonase (Pon1) Q192R polymorphism and serum Pon1 activity in diabetic patients on maintenance hemodialysis. Clin Nephrol. 2003; 60: 257–65. [DOI] [PubMed] [Google Scholar]
- 25. Ma RX, Yan SL, Yu HW, et al The association of paraoxonase 192Gln/Arg gene polymorphism with coronary heart disease Type 2 diabetes mellitus. Chinese J Diabetes. 2003; 11: 29–33. [Google Scholar]
- 26. Wang Y, Chang ZW. The association of paraoxonasel gene 192 Gln/Arg polymorphism and type 2 diabetes mellitus complicated by coronary heart disease. Guangdong Med J. 2003; 24: 598–600. [Google Scholar]
- 27. Hao YL, Li LX, Cheng G. Relation between paraoxonase 1 gene Q191R polymorphism and type 2 diabetes mellitus complicated with hypertension. Chinese J Endocrinol Metab. 2003; 19: 429–32. [Google Scholar]
- 28. Pu X, Zeng FR, Zeng ZC, et al Study on relationship between paraoxonase‐1 and type 2 diabetes mellitus in the elderly. Chinese J Geriatrics. 2003; 22: 466–8. [Google Scholar]
- 29. Ren T, Xiang KS, Liu LM. An association study of diabetic neuropathy with polymorphisms of eNOS, PON1, RAGE and ALR2 Genes. Shanghai Med J. 2003; 26: 24–7. [Google Scholar]
- 30. Deng YG, Zeng JB, Su SO. Glu‐Arg 192 polymorphism of paraoxnasel gene and type 2 diabetes with neuropathy. J Brain Nerv Dis. 2004; 12: 437–8. [Google Scholar]
- 31. Li SY, Li JY, Wei G, et al Study on the relationship between paraoxonase‐ 1 (PON‐ 1) and type 2 diabetes mellitus complicated by coronary artery disease. Tianjin Med J. 2004; 32: 18–21. [Google Scholar]
- 32. Zhang Z, Li J, Jiang C. PON1 gene polymorphism in NIDDM patients with cerebral infarction. Acta Aacademiae Med Qingdao Universitatis. 2004; 40: 336–7. [Google Scholar]
- 33. Sun YD, Sun SC, Zuo J, et al Polymorphism of paraoxonase in diabetic nephropathies. J Jilin University Medicine Edition. 2005; 31: 598–601. [Google Scholar]
- 34. Mastorikou M, Mackness M, Mackness B. Defective metabolism of oxidized phospholipid by HDL from people with type 2 diabetes. Diabetes. 2006; 55: 3099–103. [DOI] [PubMed] [Google Scholar]
- 35. Qi L, Lu Z, Dong YH. The association of paraoxonase‐1 192 Gln/Arg and paraoxonase‐2 311 Cys/Ser gene polymorphisms with macrovascular disease in type 2 diabetes mellitus patients. Chinese J Arteriosclerosis. 2007; 15: 703–7. [Google Scholar]
- 36. Shi GH, Wu NJ. Association between PON1 gene polymorphism and type 2 diabetic nephropathy. Clin Med China. 2007; 23: 134–6. [Google Scholar]
- 37. Flekac M, Škrha J, Zídková K, et al Paraoxonase 1 gene polymorphisms and enzyme activities in diabetes mellitus. Physiol Res. 2008; 57: 717–26. [DOI] [PubMed] [Google Scholar]
- 38. Irace C, Cortese C, Fiaschi E, et al The influence of PON1 192 polymorphism on endothelial function in diabetic subjects with or without hypertension. Hypertens Res. 2008; 31: 507–13. [DOI] [PubMed] [Google Scholar]
- 39. Unür M, Demirez E, Aǧaçhan B, et al The relationship of oral disturbances of diabetes mellitus patients with paraoxonase gene polymorphisms. Cell Biochem Funct. 2008; 26: 870–3. [DOI] [PubMed] [Google Scholar]
- 40. Gorshunska M, Karachentsev I, Atramentova L, et al Paraoxonase 192 Q/R gene polymorphism, enzyme activity and coronary heart disease in type 2 diabetes. Diabetologia. 2009; 52: S522–3. [Google Scholar]
- 41. Bhaskar S, Ganesan M, Chandak GR, et al Association of PON1 and APOA5 gene polymorphisms in a cohort of indian patients having coronary artery disease with and without type 2 diabetes. Genet Test Mol Biomarkers. 2011; 15: 507–12. [DOI] [PubMed] [Google Scholar]
- 42. Ergun MA, Yurtcu E, Demirci H, et al PON1 55 and 192 gene polymorphisms in type 2 diabetes mellitus patients in a Turkish population. Biochem Genet. 2011; 49: 1–8. [DOI] [PubMed] [Google Scholar]
- 43. Gupta N, Binukumar BK, Singh S, et al Serum paraoxonase‐1 (PON1) activities (PONase/AREase) and polymorphisms in patients with type 2 diabetes mellitus in a North‐West Indian population. Gene. 2011; 487: 88–95. [DOI] [PubMed] [Google Scholar]
- 44. Chen XJ, Pan SZ, Zeng J. Relationship between PON1, PON2 polymorphism and type 2 diabetic nephropathy. Med J Chinese People's Armed Police Forces. 2011; 22: 153–6. [Google Scholar]
- 45. Elnoamany MF, Dawood AA, Azmy RM, et al Paraoxonase 1 gene (Gln192‐Arg) polymorphism and the risk of coronary artery disease in type 2 diabetes mellitus. Egyptian Heart J. 2012; 64: 55–62. [Google Scholar]
- 46. Zheng YQ, Lei Y, Gong JY, et al Association of paraoxonase 1 gene polymorphisms with retinopathy in type 2 diabetes mellitus. Acta Universitatis Medicinalis Anhui. 2012; 47: 1206–8. [Google Scholar]
- 47. Gökçen S, Cengiz M, Özaydin A. Serum paraoxonase levels and PON1(192) polymorphism in type 2 diabetes mellitus patients. Gazi Med J. 2013; 24: 70–3. [Google Scholar]
- 48. Shao ZY, Gonzalez II, Lucero RO, et al Association of cholesteryl ester transfer protein genotypes with paraoxonase‐1 activity, lipid profile and oxidative stress in type 2 diabetes mellitus: A study in San Luis, Argentina. J Diabetes Investig. 2014; 6: 67–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Du WL, Song L, Dai F, et al Association between the paraoxonase 1 Q192R gene polymorphism and type 2 diabetes with periodontal disease. Chin J Gerontol. 2015; 35: 2209–12. [Google Scholar]
- 50. Agachan B, Yilmaz H, Karaali Z, et al Paraoxonase 55 and 192 polymorphism and its relationship to serum paraoxonase activity and serum lipids in Turkish patients with non‐insulin dependent diabetes mellitus. Cell Biochem Funct. 2004; 22: 163–8. [DOI] [PubMed] [Google Scholar]
- 51. Shao HQ, Zhang Y. Study on relationship between paraoxonase‐1 and type 2 diabetes mellitus complicated by coronary heart disease. Shaanxi Med J. 2006; 35: 1432–3. [Google Scholar]
- 52. Altuner D, Ates I, Suzen SH, et al The relationship of PON1 QR 192 and LM 55 polymorphisms with serum paraoxonase activities of Turkish diabetic patients. Toxicol Ind Health. 2011; 27: 873–8. [DOI] [PubMed] [Google Scholar]
- 53. Consortium HP‐AS , Abdulla MA, Ahmed I, et al Mapping human genetic diversity in Asia. Science. 2009; 326: 1541–5. [DOI] [PubMed] [Google Scholar]
- 54. Senti M, Tomas M, Anglada R, et al Interrelationship of smoking, paraoxonase activity, and leisure time physical activity: a population‐based study. Eur J Intern Med. 2003; 14: 178–84. [DOI] [PubMed] [Google Scholar]
- 55. Rao MN, Marmillot P, Gong M, et al Light, but not heavy alcohol drinking, stimulates paraoxonase by upregulating liver mRNA in rats and humans. Metab, Clin Exp. 2003; 52: 1287–94. [DOI] [PubMed] [Google Scholar]
- 56. Otocka‐Kmiecik A, Orlowska‐Majdak M. The role of genetic (PON1 polymorphism) and environmental factors, especially physical activity, in antioxidant function of paraoxonase. Postepy Hig Med Dosw. 2009; 63: 668–77. [PubMed] [Google Scholar]


