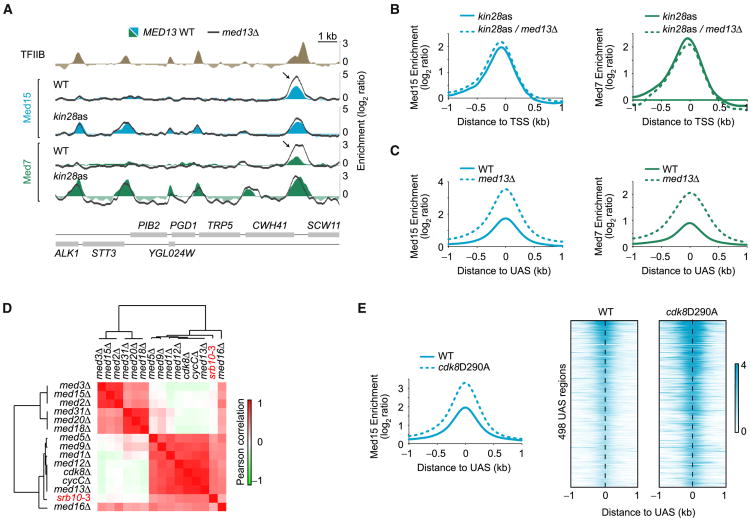
Figure 2. Mediator Accumulates at UAS Regions in the Absence of CKM.
(A) Med15 (blue) and Med7 (green) occupancy in WT and kin28as cells, all treated with NAPP1, along a segment of chromosome VII. Dark gray traces show the effect of deleting MED13 in these strains. The Med15 and Med7 peaks appearing in kin28as cells correspond to promoter peaks and are not enhanced when deleting MED13 (med13Δ, dark gray trace). Arrows are pointing toward a UAS Mediator peak, enhanced upon MED13 deletion.
(B) Average Med15 (blue) and Med7 (green) occupancy around the transcription start site (TSS) of transcriptionally active genes (n = 165, see Experimental Procedures) in kin28as cells (solid traces) and in kin28as/med13Δ cells (dashed traces). See also Figure S2A.
(C) Average Med15 (blue) and Med7 (green) enrichment around UAS regions (n = 498) in WT (solid traces) and med13Δ (dashed traces) cells.
(D) Heatmap representation of the hierarchical clustering of Pearson correlations between expression profiles of various Mediator subunit mutants. Data for srb10-3, a strain with a point mutation (D290A) in the catalytic domain of the CDK8 gene (Liao et al., 1995), were from Holstege et al. (1998), while all other data sets are from Kemmeren et al. (2014). See also Figure S2B.
(E) Average (left) and heatmap representation (right) of Med15 occupancy around UAS regions (n = 498) in WT (solid traces) and cdk8D290A (dashed traces) cells.