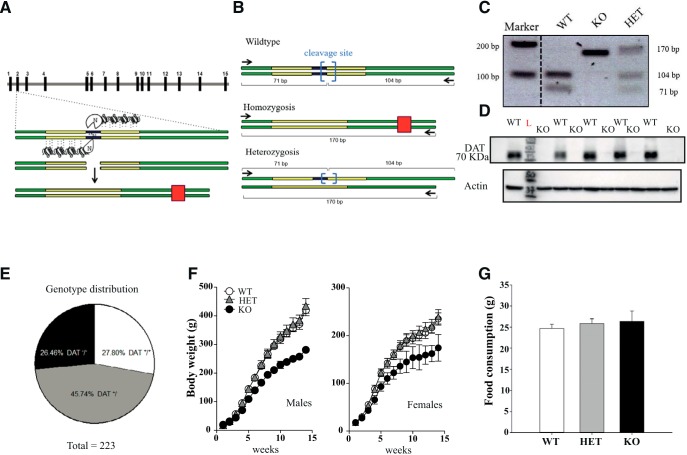
Figure 1.
Generation of DAT KO (KO) rats. A, DAT-KO rats were generated by using ZFN technology that produces a 5 bp deletion and an early stop codon. Green solid lines indicate exons 2 and 3, gray solid lines indicate introns; yellow boxes indicate DNA domains interaction sites; blue box indicates KO targeting site; red box indicates early stop codon generated after frameshift due to cleavage of 5 bp operated by ZFN. B, C, PCR primers were designed (black arrows). WT DNA contains BtsIMutI restriction enzyme site (blue brackets). WT-amplified DNA is fully digested into two low-molecular-weight bands (104 and 71 bp). Homozygote-mutated DNA loses digestion sites on both alleles, therefore resulting in only one final PCR product of 170 bp. Heterozygosis shows both digested DNA at lower molecular weight from the WT allele and mutated DNA at 170 bp. D, Western blot analysis of DAT expression in striatal samples of WT and DAT-KO rats. DAT-KO rats, in contrast to WT controls, show complete absence of DAT protein expression. Another representative blot is shown in Figure 1-1. E, Rats were bred following a HET–HET female breeding scheme. A total of 223 animals were born from 19 offspring and we obtained 62 DAT+/+ (26.46%), 102 DAT+/− (45.74%), and 59 DAT−/− (27.80%), numbers very close to the ratio (1:2:1) expected in the absence of prenatal and postnatal death. Infantile mortality was completely absent for all 3 genotypes followed for up to 4 months. F, Developmental phenotype. Both DAT-KO male and female rats develop normally but have lower weight compared with DAT-HET and WT rats (n = 10–20 per group). G, Analysis of food consumption for 3 d (ad libitum) revealed no difference between genotypes (males, n = 6–8 per genotype).