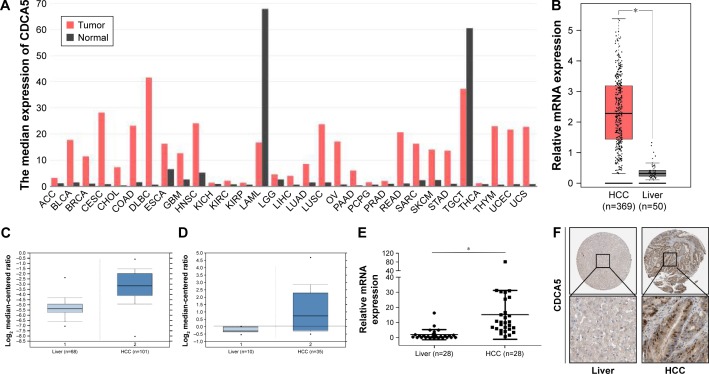
Figure 1.
CDCA5 expression is increased in human HCC specimens.
Notes: (A) CDCA5 transcription levels in various types of cancers; the graph was generated from the GEPIA website and is based on the expression of CDCA5 in various types of cancers from multiple cohorts of TCGA study. CDCA5 expression is presented as “median expression of CDCA5” in each cancer type. (B) CDCA5 expression in HCC tissues (n=369), and normal liver tissues (n=50) was analyzed based on that in the TCGA, as accessible on the GEPIA website (*P<0.05). (C) CDCA5 mRNA in HCC (n=101) and normal liver tissues (n=68) was analyzed based on that presented in Chen’s published dataset (GEO accession: GSE3500) through the Oncomine platform (P<0.05). (D) CDCA5 mRNA in HCC (n=35) and normal liver tissues (n=10) was analyzed based on that in Wurmbach’s published dataset (GEO accession: GSE6764) through the Oncomine platform (P<0.05). (E) CDCA5 expression was determined in 28 pairs of HCC samples using Q-PCR and was normalized to GAPDH (*P<0.05); HCC versus normal liver tissues. (F) CDCA5 protein expression in HCC and normal liver tissues was analyzed using the HPA website, from which the present images were retrieved. Magnification is ×40 and ×400, respectively.
Abbreviations: GEPIA, Gene Expression Profiling Interactive Analysis; HCC, hepatocellular carcinoma; HPA, Human Protein Atlas; HRs, hazard ratios; Q-PCR, quantitative polymerase chain reaction; TCGA, the Cancer Genome Atlas.