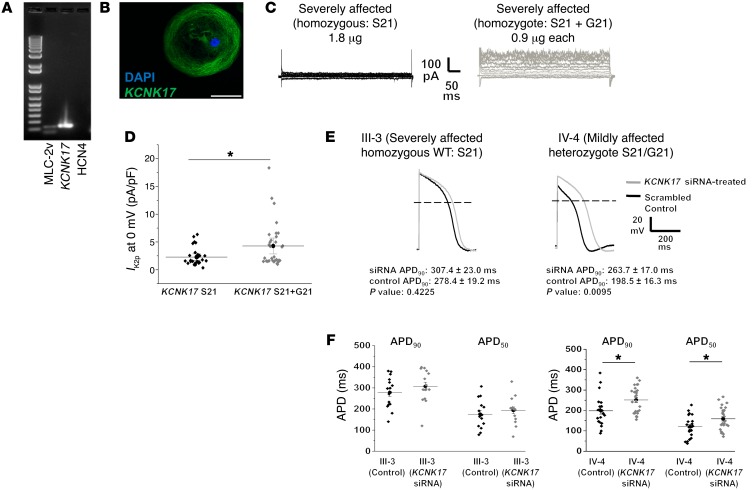
Figure 6. Two-pore potassium channel KCNK17 variant produces significant increase in K2p current.
(A) Single-cell RT-PCR confirms KCNK17 expression and ventricular lineage in electrophysiology-verified ventricular patient iPSC-CMs. MLC-2v, myosin light chain 2 ventricle (ventricular lineage marker); HCN4, hyperpolarization-activated cyclic nucleotide-gated potassium channel 4 (pacemaker lineage marker). (B) Immunohistochemistry confirms KCNK17 expression in iCell ventricular cardiomyocyte. Blue, nucleus; green, KCNK17. Scale bar: 50 μm. (C) Representative macroscopic current-voltage traces obtained from homozygous WT (KCNK17 S21 only, mimics severely affected patient alleles) and heterozygous state (KCNK17 S21 + G21, mimics mildly affected patient alleles). (D) Mean current density analyzed at 0 mV between WT and heterozygote conditions from 3 independent experiments. Relative current KCNK17 S21: 2.27 ± 0.30 (n = 29); and KCNK17 S21 + G21: 4.28 ± 0.70 (n = 32). *P = 0.01 as determined by unpaired Student’s t test. Results normalized to capacitance and shown as mean ± SEM. (E) Representative macroscopic action potential traces following KCNK17 siRNA silencing (gray) compared with scrambled control (black) in III-3 (P = 0.42, n = 15 in each group) and IV-4 (P = 0.01, n = 21 in control, n = 27 in siRNA) as determined by unpaired Student’s t test. (F) Summary APD90 and APD50 data from III-3 and IV-4 in control versus KCNK17 siRNA groups. *P < 0.02 as determined by unpaired Student’s t test. No significant changes were observed in mean diastolic potential (MDP) or action potential amplitude (APA) between siRNA and control recordings from III-3. MDP was unchanged in IV-4, but APA was higher in the siRNA group (100.26 ± 2.73 mV) compared with control (87.24 ± 2.18 mV) (Supplemental Table 6).