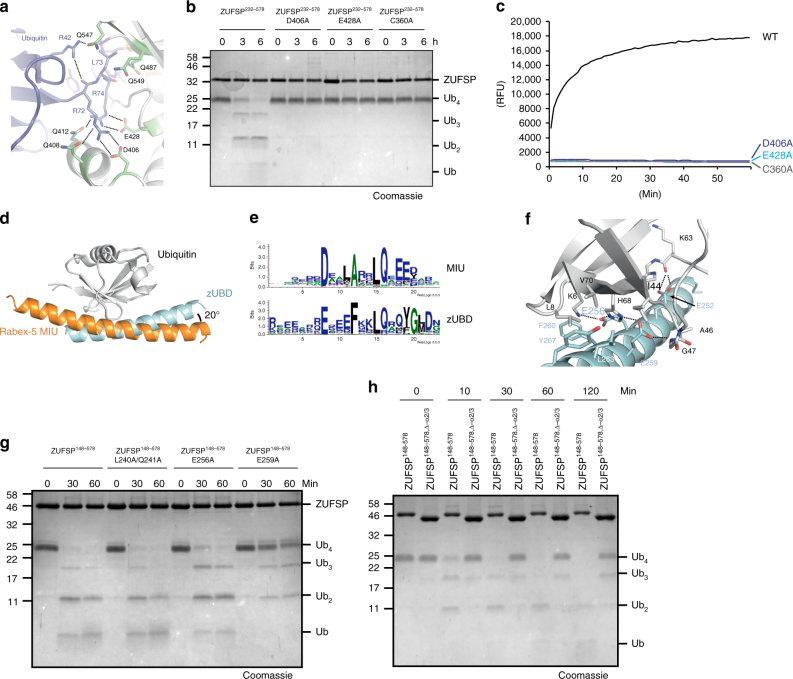
Fig. 5.
Determinants of chain specificity. a Recognition of ubiquitin C terminus by the catalytic core of ZUFSP. Ubiquitin (blue) and ZUFSP (gray/green) shown in cartoon representation with key residues highlighted as sticks. Blue and black residue labels refer to ubiquitin and ZUFSP, respectively. Salt bridges are indicated by dotted lines. b Activity of C-terminus recognition mutants (ZUFSP232–578 D406A or E428A) against K63-linked Ub4 was compared to ZUFSP232–578 and inactive ZUFSP (ZUFSP232–578 C360A). c Activity of mutants described in b against RLRGG-AMC. The RFU values shown are the means of triplicates. d Structural superposition of ubiquitin-binding interfaces to zUBD (cyan, this work) and the MIU domain of Rabex-5 (2FIF, orange). Orientation of the two helical ubiquitin-binding domains differ by 20°. e SeqLogo45 representation of the consensus sequences for the MIU motif14 (top) and the zUBD derived from the ZUFSP family as shown in Supplementary Fig. 1 (bottom). f Magnification of the interaction interface between ubiquitin and zUBD. Relevant residues are shown as sticks and labeled black in case of ubiquitin and cyan in case of zUBD. Electrostatic interactions are indicated as dotted lines. g Activity of a MIU mutant (ZUFSP148–578 L240A/Q241A) and two zUBD mutants (ZUFSP148–578E256A and E259A) on K63-linked Ub4, in comparison to wild-type ZUFSP148-578. h Activity time course of the α2/3-deletion mutant ZUFSP148–578; Δ-α2/3 on K63-linked Ub4 chains, compared to activity of the parental ZUFSP148–578 construct