ABSTRACT
The nuclear pore complex (NPC) is the sole gateway for molecular transport between the nucleus and the cytoplasm in eukaryotes. The NPC is composed of approximately 30 different kinds of protein components called nucleoporins. The functional structure of the NPC is highly conserved among various eukaryotes. However, the exact mechanisms by which many of the nucleoporins exert their specific functions still remain unclear. The single cell ciliate Tetrahymena has two functionally distinct nuclei, a macronucleus and a micronucleus, and we have discovered that these two nuclei have compositionally distinct NPCs. We initially identified four nucleus-specific Nup98-s and demonstrated that they are required for macronucleus- and micronucleus-specific nuclear transport. More recently we identified two nucleus-specific paralogs of Nup214 and Nup153 and two nucleus-specific transmembrane proteins Pom121 and Pom82. Our findings argue that Nup214, Nup153, and Pom121/Pom82 also act as key molecules for transport machineries to differentiate the two functionally distinct nuclei present in this organism.
KEYWORDS: nuclear pore complex; nucleoporin; macronucleus, micronucleus; ciliate; Tetrahymena thermophila
The nucleus of the eukaryotic cell is compartmentalized from the cytoplasm by the nuclear envelope (NE). Nuclear pores present in the NE allow for molecular transport between cytoplasm and nucleus. These nuclear pores are formed by a nuclear pore complex (NPC), a huge protein complex approximately 60 MDa in yeast and ∼120 MDa in mammalian cells, composed of multicopies of around 30 kinds of protein species called nucleoporins (Nups).1–3
As a ciliated protozoan, Tetrahymena thermophila, a unicellular eukaryote, has dimorphic nuclei that are functionally distinct, a highly expressed somatic macronucleus (MAC)17 and a transcriptionally silent germline micronucleus (MIC),18 in a single cytoplasm (Fig. 1A). These nuclei replicate their DNA at different times and the method of nuclear division is different: an amitosis segregates centromere-lacking MAC chromosomes whereas conventional mitosis segregates MIC chromosomes.19 These facts are evidence that the two nuclei communicate independently with the cytoplasm and that functionally distinct NPCs are likely required by each nucleus.
Figure 1.
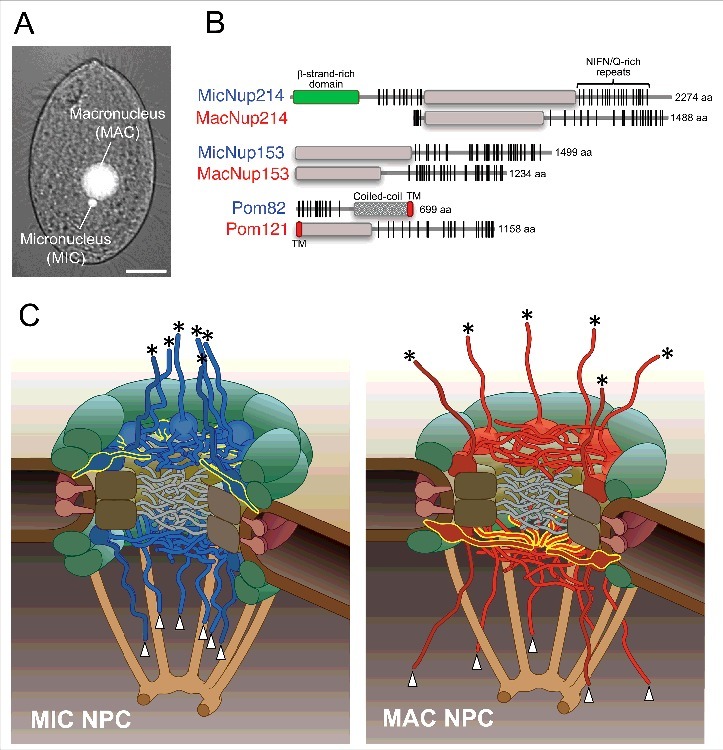
(A) A bright field micrograph of T. thermophila overlaid with fluorescence images of the macronucleus (MAC) and micronucleus (MIC) stained with a DNA-specific fluorescent dye, 4′,6- diamidino-2-phenylindole (DAPI). Scale bar, 10 μm. (B) Molecular domain structures of nucleus-specific FG-Nups. Blue or red letters in the protein names denote MIC-specific or MAC-specific components, respectively. The green box is a β-strand-rich domain. Gray boxes are ordered regions mainly composed of α-helices. Red ellipses are predicted transmembrane helices. Vertical lines indicate the positions of FG or FX repeats (X means any residues, but the majority are N and Q). (C) The structural model of Tetrahymena NPCs. Left panel is a MIC NPC and the right panel is a MAC NPC. Upper and lower sides are cytoplasmic and nucleoplasmic sides, respectively. The components represented as filamentous structure are FG-Nups. Red and blue indicate MAC- and MIC-specific FG-Nups, respectively. FG-Nups extending to the cytoplasmic side indicated by asterisks are MicNup214 (in the left panel) and MacNup214 (in the right panel). The components extending to nucleoplasmic side indicated by arrowheads are MicNup153 (in the left panel) and MacNup153 (in the right panel). Pom82 in the MIC NPC and Pom121 in the MAC NPC are distinguished by yellow outlines. Central channel FG-Nups that are shared by both NPCs are colored in gray. Green and brown blocks are Nup107-160 complexes (Y-complex) and Nup93 complexes, respectively.
Structural components of the eukaryotic NPC
The Nups can be classified into three categories by their molecular structures and functions. The first group of nucleoporins are transmembrane proteins bearing an α-helix domain. These transmembrane Nups penetrate the NE and anchor the NPC to the pore membrane.
The second group of nucleoporins are scaffold proteins bearing both α-helices and β-strands over their entire length. These proteins compose the framework of the NPC. There are two well defined subcomplexes of the proteins categorized into this group: one is the Nup93 complex (the Nic96 complex in budding yeast) forming the inner ring structure of the NPC,4,5 and the other is the Nup107-160 complex (the Nup84-120 complex in budding yeast, also called the Y-complex)6,7 forming nuclear and cytoplasmic outer ring structures.8,9
The last group of nucleoporins are the FG-repeat proteins bearing repeats of Phe-Gly (FG) resides that have an unfolded conformation.10 In the NPC, the FG repeat regions have a hydrophobic nature and form an entropic barrier that functions to exclude nonspecific passage of molecules over approximately 40 kDa, but facilitates the passage of nuclear transport receptors (NTRs). There are several models under debate of how FG-repeat regions form an entropic barrier, and also how the NTRs pass through the pore by interacting with this entropic barrier.11-16
Tetrahymena MAC and MIC NPCs
The NPCs of the MAC and MIC of T. thermophila are remarkably distinctive in the signature of the FG-repeat regions of the Nup98 orthologs: the MAC-specific MacNup98A and B have Gly-Leu-Phe-Gly (GLFG) repeats and the MIC-specific MicNup98A and B have Asn-Ile-Phe-Asn (NIFN) repeats.20,21 These distinct Nup98 repeats function in exclusive nuclear transport of nucleus-specific linker histones,20 suggesting that functional differentiation of the NPCs based on distinct structures of selective Nups is related to the different functions of the nuclei.20-22
Our comprehensive search for T. thermophila nucleoporins identified a total of 28 proteins.20,23 Characterization of these nucleoporins clearly showed that there are nucleus-specific Nups other than the Nup98s: MicNup214, MicNup153, and Pom82 are MIC-specific, and MacNup214, MacNup153, and Pom121 are MAC-specific (Fig. 1B). The other Nups, including most FG-Nups that locate to the central channel and the structural Nups that are involved in assembling the Nup93- and Y-complexes are shared by the two nuclei. However, there is a difference in the number of Y-complexes of the two nuclei: there are more Y-complexes in the MIC NPC than the MAC NPC (see green components in Fig. 1C), although, the effects of this difference are currently unknown. The other components seem to be equally localized to both NPCs (see grey and brown components in Fig. 1C). Importantly, all of the nucleus-specific Nups, including the transmembrane Pom82 and Pom121 (see yellow-outlined components in Fig. 1C), are FG-repeat containing Nups that directly function in nuclear transport, strongly suggesting that the differences in T. thermophila MAC and MIC NPCs result in specialized nuclear transport for each nucleus.
Possible functions of nucleus-specific Nup214, Nup153 and Pom121/pom82
It is known that in other organisms that Nup214 and Nup153 are localized to the cytoplasmic and nucleoplasmic sides of the NPC, respectively.24,25 These asymmetrical FG-Nups are probably involved in the selection of nuclear transport cargos at the entrance and release at the exit of the NPC. Similarly to Nup98, MicNup214 and MacNup214 are clearly different in their repeat signatures. MicNup214 has a NIFN/Q-rich repeat while MacNup214 has a more conventional FG repeat signature (Fig. 1B). Therefore, Nup214s may function in the regulation of nucleus-specific transport in cooperation with nucleus-specific Nup98s. In addition, the β-strand-rich structure in the N-terminal region that is generally conserved in the Nup214 homologs is present in MicNup214, but is lacking in MacNup214 (Fig. 1B). This β-strand-rich domain is known to interact with DEAD-box RNA helicases in yeast and human cells.26,27 Although the function of the β-strand-rich domain of Tetrahymena MicNup214 has not been experimentally determined, the presence or absence of this structure may be involved in a divergence of nuclear function.
In contrast to Nup214s, MicNup153 and MacNup153 don't have significant differences in their FG-repeat signatures,23 therefore, they may not have intrinsic differences in nuclear transport function. However, the N-terminal structural domains of the two proteins, which by analogy with human Nup15328 are presumed to have Ran binding activity as well as NPC targeting activity, are somewhat different. Ran, a GTPase, is a key regulator of nuclear transport, and differences in Ran binding by MicNup153 and MacNup153 could provide significant differences in their nuclear transport functions and control the release of cargo at the NPC in a nucleus-specific manner. Currently, however, the Ran binding properties of the Nup153s have not yet been examined. Beside its nuclear transport functions, Nup153 is known to be involved in the regulation of gene expression in mammalian cells,29 suggesting that in Tetrahymena MacNup153 may have a role in the regulation of gene expression. Because no gene expression occurs in the MIC during vegetative growth, MicNup153 does not have an active role on gene expression in vegetative cells. Further studies are required for understanding the functions of the nucleus-specific Nup153-s.
Both MAC-specific Pom121 and MIC-specific Pom82 are transmembrane Nups bearing FG-repeats (Fig. 1B). It was surprising that Tetrahymena had a Pom121-like nucleoporin, because Pom121 has so far been found only in vertebrate species30; although, it is unclear whether vertebrate and Tetrahymena Pom121 have a common origin of molecular evolution. The localization of Pom121 is biased to the nucleoplasmic face of the MAC NPC. Pom82, in contrast, is localized to the cytoplasmic face of the MIC NPC, and it exhibits a decidedly different organization of secondary structures and domains from those of Pom121 (Fig. 1B). These differences imply that Pom82 is functionally and evolutionary different from Pom121. Pom82 may be a Tetrahymena-specific Nup, because no Nups with similar molecular architecture have so far been found in other organisms including Paramecium, Oxytricha, and Stentor, although, functional homologs may exist in those species. The fact that Pom121 and Pom82 are nucleus-specific suggests that the targeting of these transmembrane proteins to the MAC and MIC is an initial step in the assembly of compositionally different NPCs in the same cytoplasm. To confirm this hypothesis, it will need to be determined which Nups associate with these Pom proteins within the NPC architecture. Resolving when and how Tetrahymena acquired these Pom proteins during their evolution may help in understanding the establishment of ciliate nuclear dimorphism.
Nuclear dimorphism is one of the most important issues in ciliate biology. Because the nucleus-specific Nups can be easily distinguished by transport factors, these Nups can function as key factors in various nucleus-specific events, including nucleus-specific control of cell cycle progression during cell proliferation and nuclear differentiation during sexual reproduction. Future studies on the dynamic behavior and functions of these nucleus-specific Nups will open new windows to understanding nuclear dimorphism in ciliates.
Acknowledgments
We thank Dr. David B. Alexander for critical reading of the manuscript.
Funding
This work was supported by grants from JST to T. H., Uehara Memorial Foundation to T. H. and from JSPS KAKENHI Grant Numbers JP24570227, JP15K07066 to M. I., JP17H01444, JP16H01309 to Y. H. and JP23114724, JP25116006, JP26291007, JP17H03636 to T. H.
References
- 1.Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao Y, Chait BT. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J Cell Biol. 2000;148:635–51. doi: 10.1083/jcb.148.4.635. PMID:10684247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cronshaw JM, Krutchinsky AN, Zhang W, Chait BT, Matunis MJ. Proteomic analysis of the mammalian nuclear pore complex. J Cell Biol. 2002;158:915–27. doi: 10.1083/jcb.200206106. PMID:12196509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raices M, D'Angelo MA. Nuclear pore complex composition: a new regulator of tissue-specific and developmental functions. Nat Rev Mol Cell Biol. 2012;13:687–99. doi: 10.1038/nrm3461. PMID:23090414. [DOI] [PubMed] [Google Scholar]
- 4.Stuwe T, Bley CJ, Thierbach K, Petrovic S, Schilbach S, Mayo DJ, Perriches T, Rundlet EJ, Jeon YE, Collins LN, et al.. Architecture of the fungal nuclear pore inner ring complex. Science. 2015;350:56–64. doi: 10.1126/science.aac9176. PMID:26316600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kosinski J, Mosalaganti S, von Appen A, Teimer R, DiGuilio AL, Wan W, Bui KH, Hagen WJ, Briggs JA, Glavy JS, et al.. Molecular architecture of the inner ring scaffold of the human nuclear pore complex. Science. 2016;352:363–5. doi: 10.1126/science.aaf0643. PMID:27081072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Siniossoglou S, Lutzmann M, Santos-Rosa H, Leonard K, Mueller S, Aebi U, Hurt E. Structure and assembly of the Nup84p complex. J Cell Biol. 2000;149:41–54. doi: 10.1083/jcb.149.1.41. PMID:10747086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walther TC, Alves A, Pickersgill H, Loïodice I, Hetzer M, Galy V, Hülsmann BB, Köcher T, Wilm M, Allen T, et al.. The conserved Nup107-160 complex is critical for nuclear pore complex assembly. Cell. 2003;113:195–206. doi: 10.1016/S0092-8674(03)00235-6. PMID:12705868. [DOI] [PubMed] [Google Scholar]
- 8.Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al.. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. doi: 10.1038/nature06405. PMID:18046406. [DOI] [PubMed] [Google Scholar]
- 9.von Appen A Kosinski J, Sparks L, Ori A, DiGuilio AL, Vollmer B, Mackmull MT, Banterle N, Parca L, Kastritis P, et al.. In situ structural analysis of the human nuclear pore complex. Nature. 2015;526:140–3. doi: 10.1038/nature15381. PMID:26416747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Terry LJ, Wente SR. Flexible gates: dynamic topologies and functions for FG nucleoporins in nucleocytoplasmic transport. Eukaryot Cell. 2009;8:1814–27. doi: 10.1128/EC.00225-09. PMID:19801417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ribbeck K, Görlich D. Kinetic analysis of translocation through nuclear pore complexes. EMBO J. 2001;20:1320–30. doi: 10.1093/emboj/20.6.1320. PMID:11250898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rout MP, Aitchison JD, Magnasco MO, Chait BT. Virtual gating and nuclear transport: the hole picture. Trends Cell Biol. 2003;13:622–8. doi: 10.1016/j.tcb.2003.10.007. PMID:14624840. [DOI] [PubMed] [Google Scholar]
- 13.Frey S, Görlich D. A saturated FG-repeat hydrogel can reproduce the permeability properties of nuclear pore complexes. Cell. 2007;130:512–23. doi: 10.1016/j.cell.2007.06.024. PMID:17693259. [DOI] [PubMed] [Google Scholar]
- 14.Patel SS, Belmont BJ, Sante JM, Rexach MF. Natively unfolded nucleoporins gate protein diffusion across the nuclear pore complex. Cell. 2007;129:83–96. doi: 10.1016/j.cell.2007.01.044. PMID:17418788. [DOI] [PubMed] [Google Scholar]
- 15.Lim RY, Fahrenkrog B, Köser J, Schwarz-Herion K, Deng J, Aebi U. Nanomechanical basis of selective gating by the nuclear pore complex. Science. 2007;318:640–3. doi: 10.1126/science.1145980. PMID:17916694. [DOI] [PubMed] [Google Scholar]
- 16.Yamada J, Phillips JL, Patel S, Goldfien G, Calestagne-Morelli A, Huang H, Reza R, Acheson J, Krishnan VV, Newsam S, et al.. A bimodal distribution of two distinct categories of intrinsically disordered structures with separate functions in FG nucleoporins. Mol Cell Proteomics. 2010;9:2205–24. doi: 10.1074/mcp.M000035-MCP201. PMID:20368288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eisen JA, Coyne RS, Wu M, Wu D, Thiagarajan M, Wortman JR, Badger JH, Ren Q, Amedeo P, Jones KM, et al.. Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLOS Biol. 2006;4:e286. doi: 10.1371/journal.pbio.0040286. PMID:16933976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamilton EP, Kapusta A, Huvos PE, Bidwell SL, Zafar N, Tang H, Hadjithomas M, Krishnakumar V, Badger JH, Caler EV, et al.. Structure of the germline genome of Tetrahymena thermophila and relationship to the massively rearranged somatic genome. eLife. 2016;5:e19090. doi: 10.7554/eLife.19090. PMID:27892853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Orias E, Cervantes MD, Hamilton EP. Tetrahymena thermophila, a unicellular eukaryote with separate germline and somatic genomes. Res Microbiol. 2011;162:578–86. doi: 10.1016/j.resmic.2011.05.001. PMID:21624459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iwamoto M, Mori C, Kojidani T, Bunai F, Hori T, Fukagawa T, Hiraoka Y, Haraguchi T. Two distinct repeat sequences of Nup98 nucleoporins characterize dual nuclei in the binucleated ciliate Tetrahymena. Curr Biol. 2009;19:843–7. doi: 10.1016/j.cub.2009.03.055. PMID:19375312. [DOI] [PubMed] [Google Scholar]
- 21.Iwamoto M, Asakawa H, Hiraoka Y, Haraguchi T. Nucleoporin Nup98: a gatekeeper in the eukaryotic kingdoms. Genes Cells. 2010;15:661–9. doi: 10.1111/j.1365-2443.2010.01415.x. PMID:20545767. [DOI] [PubMed] [Google Scholar]
- 22.Iwamoto M, Hiraoka Y, Haraguchi T. The nuclear pore complex acts as a master switch for nuclear differentiation. Commun Integr Biol. 2015;8:4, e1056950: doi: 10.1080/19420889.2015.1056950. PMID:26479399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iwamoto M, Osakada H, Mori C, Fukuda Y, Nagao K, Obuse C, Hiraoka Y, Haraguchi T. Compositionally distinct nuclear pore complexes of functionally distinct dimorphic nuclei in ciliate Tetrahymena. J Cell Sci. 2017;130:1822–34. doi: 10.1242/jcs.199398. PMID:28386019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kraemer D, Wozniak RW, Blobel G, Radu A. The human CAN protein, a putative oncogene product associated with myeloid leukemogenesis, is a nuclear pore complex protein that faces the cytoplasm. Proc Natl Acad Sci U S A 1994;91:1519–23. doi: 10.1073/pnas.91.4.1519. PMID:8108440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sukegawa J, Blobel G. A nuclear pore complex protein that contains zinc finger motifs, binds DNA, and faces the nucleoplasm. Cell 1993;72:29–38. doi: 10.1016/0092-8674(93)90047-T. PMID:8422679. [DOI] [PubMed] [Google Scholar]
- 26.Schmitt C, von Kobbe C, Bachi A, Panté N, Rodrigues JP, Boscheron C, Rigaut G, Wilm M, Séraphin B, Carmo-Fonseca M, et al.. Dbp5, a DEAD-box protein required for mRNA export, is recruited to the cytoplasmic fibrils of nuclear pore complex via a conserved interaction with CAN/Nup159p. EMBO J 1999;18:4332–47; doi: 10.1093/emboj/18.15.4332. PMID:10428971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Napetschnig J, Kassube SA, Debler EW, Wong RW, Blobel G, Hoelz A. Structural and functional analysis of the interaction between the nucleoporin Nup214 and the DEAD-box helicase Ddx19. Proc Natl Acad Sci U S A. 2009;106:3089–94; doi: 10.1073/pnas.0813267106. PMID:19208808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakielny S, Shaikh S, Burke B, Dreyfuss G. Nup153 is an M9-containing mobile nucleoporin with a novel Ran-binding domain. EMBO J. 1999;18:1982–95. doi: 10.1093/emboj/18.7.1982. PMID:10202161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ibarra A, Benner C, Tyagi S, Cool J, Hetzer MW. Nucleoporin-mediated regulation of cell identity genes. Genes Dev. 2016;30:2253–8. doi: 10.1101/gad.287417.116. PMID:27807035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Field MC, Koreny L, Rout MP. Enriching the pore: splendid complexity from humble origins. Traffic. 2014;15:141–56. doi: 10.1111/tra.12141. PMID:24279500. [DOI] [PMC free article] [PubMed] [Google Scholar]


