Figure 3.
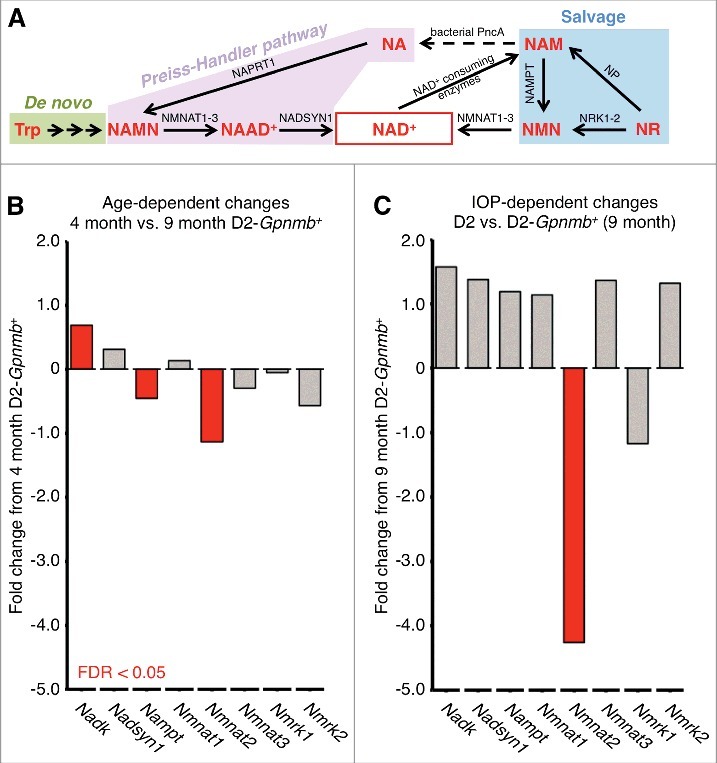
NAD synthesis and NAD relevant genes in RGCs. (A) NAD synthesis. Tryptophan (Trp) is used to form NAD+ de novo from diet in an 8 step pathway with nicotinic acid mononucleotide (NAMN) and nicotinic acid adenine dinucleotide (NAAD+) intermediates. Alternatively NAD+ can be produced through 2 other core pathways; the Preiss-Handler pathway from nicotinic acid (NA), or through the salvage pathway from nicotinamide (NAM). NA is used in the Preiss-Handler pathway to form NAD+ via 2 steps shared with the de novo pathway: NAMN (by nicotinic acid phosphoribosyltransferase; NAPRT1) and NAAD+ (by NAD+ synthetase; NADSYN1). In the salvage pathway, NAM is used to form NAD+ being converted by nicotinamide phosphoribosyltransferase (NAMPT) to nicotinamide mononucleotide (NMN) and subsequently to NAD+ by nicotinamide nucleotide adenylytransferase (NMNAT1, −2, and −3). NAM can also be converted to NA in the gut by bacterial PncA (nicotinamidase) and salvaged into the Preiss-Handler pathway. NAM is available in diet, but can also be produced by NAD+-consuming enzymes. Nicotinamide riboside (NR) can feed into the salvage pathway to form NAD+ by nicotinamide riboside kinases (NRK1, −2; Nmrk1, −2 as mouse genes) via NMN, or via NAM by purine nucleoside phosphorylase (NP). (B) and (C) Retinal ganglion cells exhibit age-dependent changes in NAD+ synthesis-related genes as well as further IOP-dependent declines in Nmnat2, an important NAD producing enzyme linked to axon protection. The decline in NAD is a major age-dependent risk factor for DBA/2J glaucoma. NADK (Nadk gene) is a major NAD-consuming kinase and its upregulation suggests increased NAD consumption / utilization. Differentially expressed genes (FDR < 0.05) are shown in red. Non-differentially expressed genes are shown in gray.
