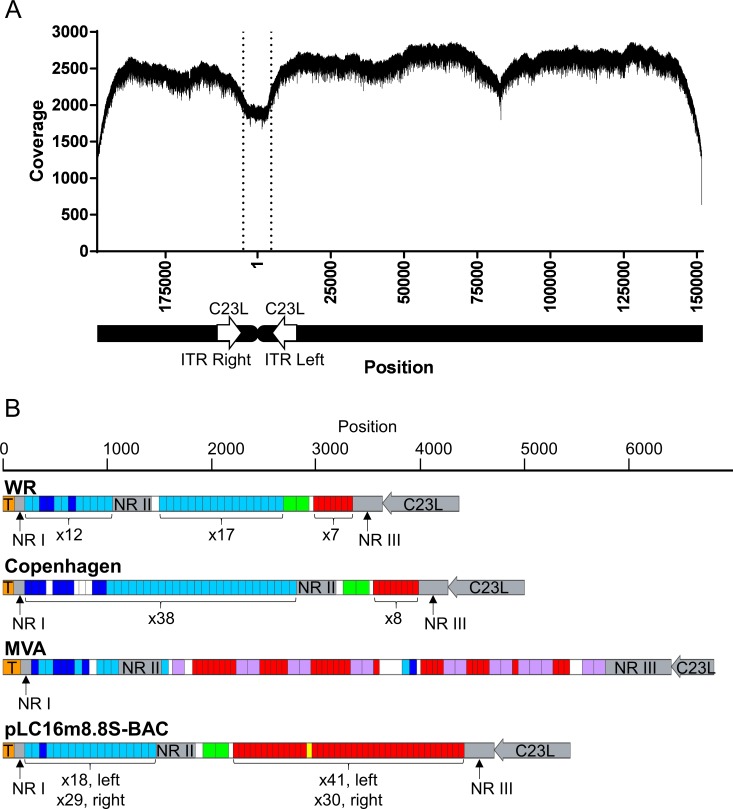
Fig 8.
Coverage depth of the pLC16m8.8S-BAC aligned to the consensus sequence of pLC16m8.8S-BAC generated by de novo assembly using HGAP software (A). The ITR regions were not arranged at the termini of the sequence for the purpose of accurate reference-based alignment. The ITR left to the right region flanked by the C23L genes is indicated by vertical dotted lines. Nucleotide position 1 is at the terminus of the ITR left. The structure of the ITR left region to the C23L gene of pLC16m8.8S-BAC (accession no. LC315596), VAC strain WR (accession no. AY243312), Copenhagen (accession no. M35027), and MVA (accession no. U94848) are shown (B). Gray rectangles indicate unique sequences in the ITR: non-repeated region (NR) I, II, III, and the C23L gene. The T in the ocher rectangle indicates a terminal hairpin. Dark or light blue rectangles correspond to 69 or 70 base pair tandem repeats. Red and yellow rectangles correspond to 53 or 54 base pair tandem repeats. Green rectangles correspond to 125 base pair repeats. Purple rectangles correspond 111 or 112 base pair repeats. The number of repeats in the left or right ITR is indicated below the repeats.