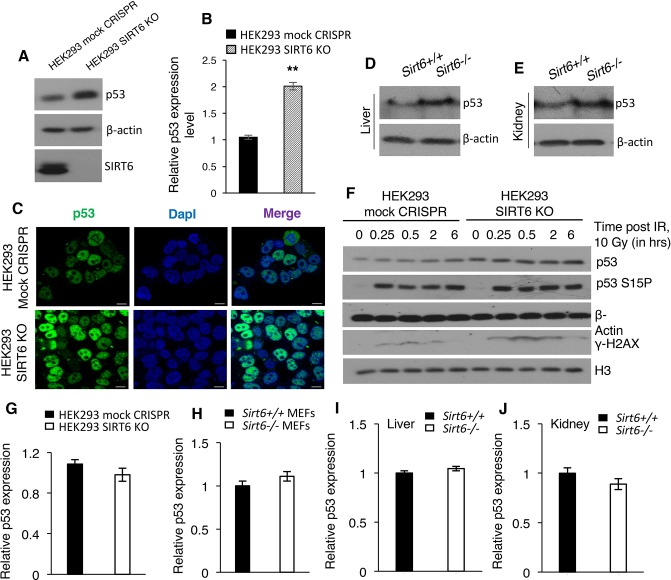
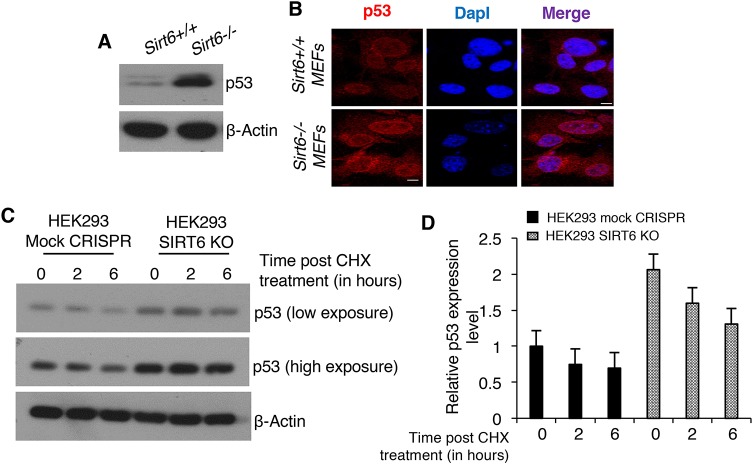
Figure 6. SIRT6 negatively regulates the stability of p53.
(A) Western blotting analysis of p53 protein expression in SIRT6 KO and control HEK293 cells. (B) Quantification of data presented in (A). Data represent mean ± SEM, n = 3. **p<0.01 calculated using Student’s t-test. (C) Immunofluorescence staining to confirm enhanced p53 expression in HEK293 mock CRISPR and SIRT6 KO cells. Scale bar, 10 µm. (D) Western blotting analysis of p53 protein expression in liver of wild-type (WT) and Sirt6-/- mice. (E) Western blotting analysis of p53 protein expression in kidneys of wild-type (WT) and Sirt6-/- mice. (F) Western blotting analysis of p53 and phosphorylation of p53 at serine 15 in HEK293 mock CRISPR and SIRT6 KO cells in response to 10 Gy of γ-irradiation. (G) qPCR analysis of p53 expression in HEK293 mock CRISPR and SIRT6 KO cells (with respect to Gapdh controls). Data represent mean ± SEM, n = 3. (H) qPCR analysis of p53 expression in Sirt6+/+ and Sirt6-/- MEFs (with respect to Gapdh controls). Data represent mean ± SEM, n = 3. (I) qPCR analysis of p53 expression in liver of Sirt6+/+ and Sirt6-/- mice (with respect to Gapdh controls). Data represent mean ± SEM, n = 3. (J) qPCR analysis of p53 expression in kidneys of Sirt6+/+ and Sirt6-/- mice (with respect to Gapdh controls). Data represent mean ± SEM, n = 3.