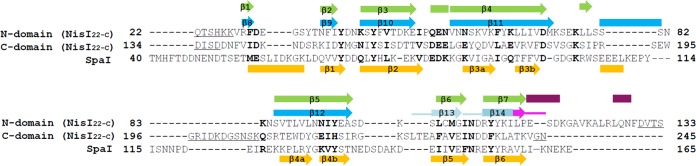
FIG 1.
Structure-guided sequence alignment. Structure-guided sequence alignment of the N- and C-terminal domains of NisI22-C from L. lactis with those of SpaI from B. subtilis. Identical and similar residues are shown in bold, gaps are shown with hyphens, and internal disordered residues are underlined. The residue numbers are relative to those of the methionine residues in the prelipoproteins before processing of the N-terminal leader sequence. Secondary structures (β-strand, arrows; α-helix, rectangles) of NisI22-C are shown above the alignment and colored in the scheme as in Fig. 2A, and those of SpaI are shown in orange below the alignment. For the 21-amino-acid fragment of the C-terminal domain of NisI22-C, which includes the 5-amino-acid C-terminal fragment (pink), the loops of the secondary structures are also shown with thick lines. The numbers of the secondary structures are assigned only to β-strands comprising the β-barrel structure, and those of SpaI are based on the published report (29).