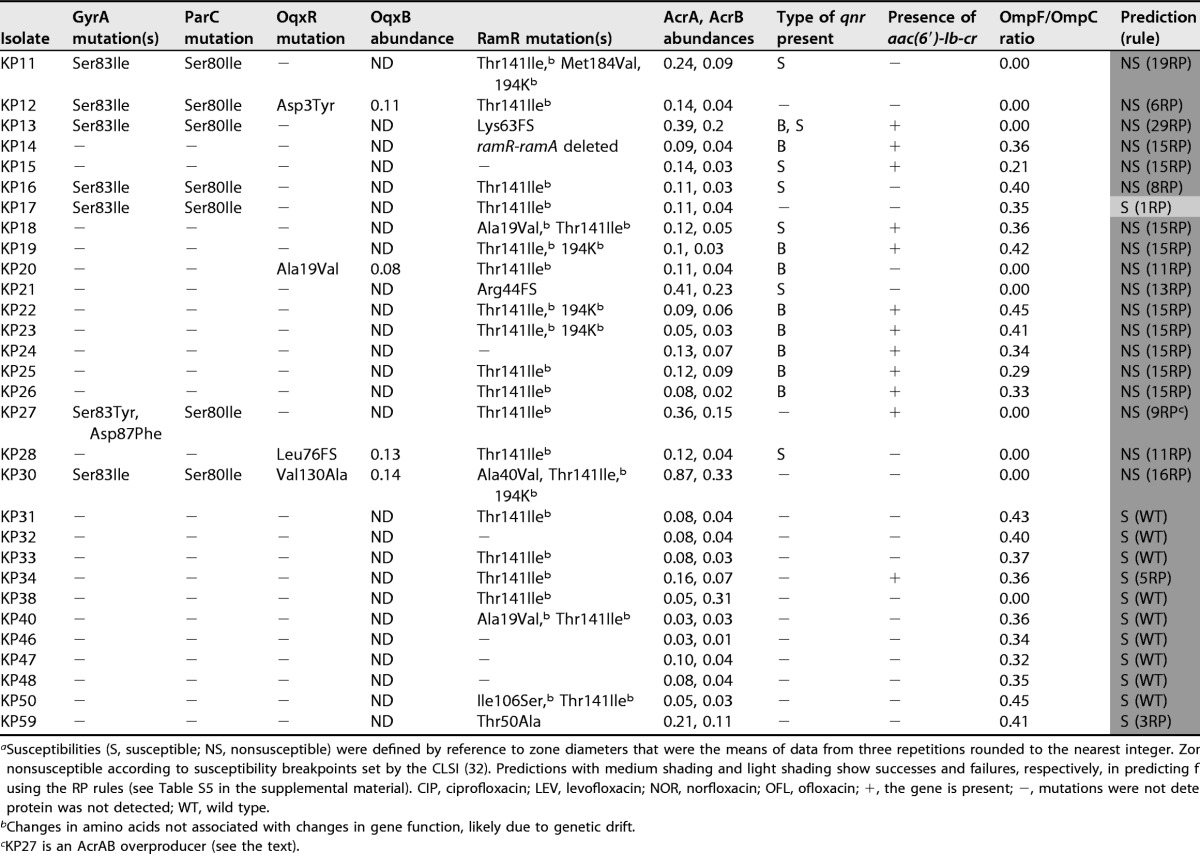
TABLE 2.
Combination of disc test, proteomics, and genome sequencing data and predictions of fluoroquinolone susceptibility for K. pneumoniae clinical isolatesa
Susceptibilities (S, susceptible; NS, nonsusceptible) were defined by reference to zone diameters that were the means of data from three repetitions rounded to the nearest integer. Zones with dark shading are nonsusceptible according to susceptibility breakpoints set by the CLSI (32). Predictions with medium shading and light shading show successes and failures, respectively, in predicting fluoroquinolone susceptibility by using the RP rules (see Table S5 in the supplemental material). CIP, ciprofloxacin; LEV, levofloxacin; NOR, norfloxacin; OFL, ofloxacin; +, the gene is present; −, mutations were not detected or the gene is absent; ND, protein was not detected; WT, wild type.
Changes in amino acids not associated with changes in gene function, likely due to genetic drift.
KP27 is an AcrAB overproducer (see the text).