Figure 2. Validation of Dam chromatin accessibility profiling compared to ATAC and FAIRE-seq.
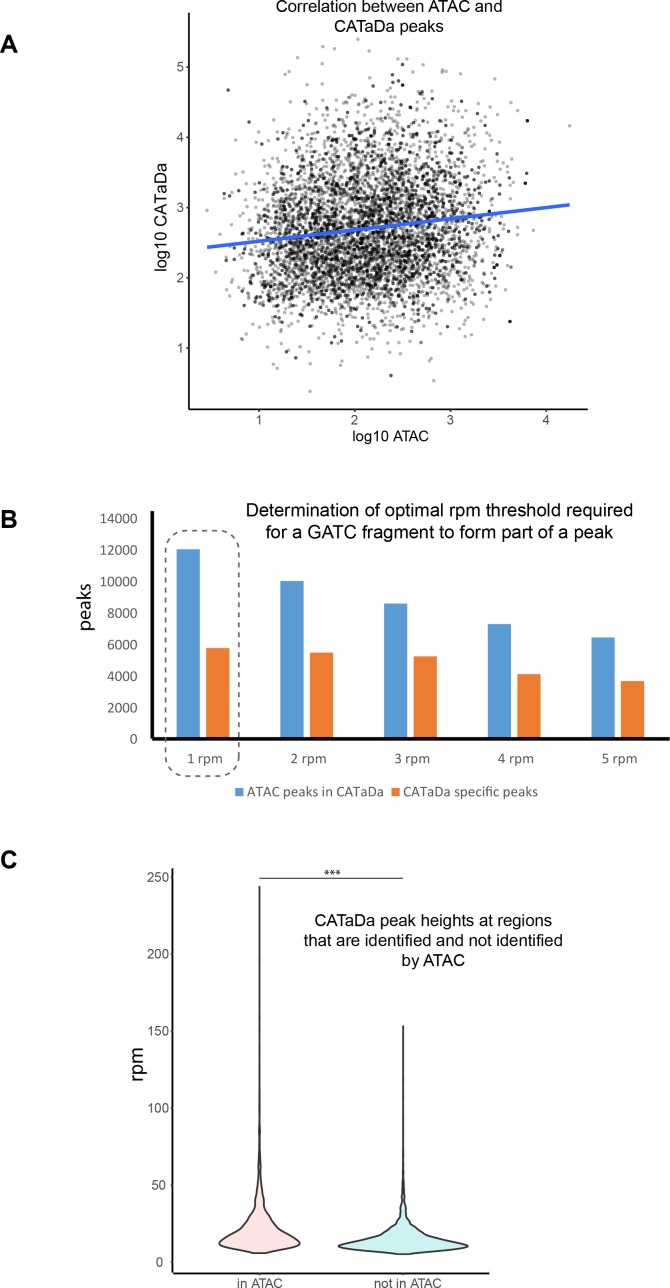
(A) Chromatin accessibility across chromosome three as determined by ATAC-seq, FAIRE-seq, and CATaDa. Note the reduced amount open chromatin proximal to the centromere regions in all three datasets. y-axes = reads per million (rpm). (B) Example locus showing data obtained by FAIRE, ATAC, and CATaDa. Peaks are broadly reproducible across techniques. (C) Aggregation plot of CATaDa signal at TSS with 2 kb regions up and downstream. Aggregated signal at TSS shows expected enrichment of Dam. (D) Aggregation plot of CATaDa signal at ATAC or FAIRE peaks, indicating enrichment of CATaDa signal at these loci. (E) Identification of ATAC peaks in CATaDa or FAIRE data. CATaDa and FAIRE identify 48.6% and 55.9% of ATAC peaks, respectively. FAIRE-seq peaks overlap more frequently with promoter proximal peaks (2 kb from TSS), whilst CATaDa peaks overlap with more ATAC peaks outside of promoter regions.