Abstract
Salt-resistant yeast strains are highly demanded by industry due to the exposure of yeast cells to high concentrations of salt, in various industrial bioprocesses. The aim of this study was to perform a physiological and transcriptomic analysis of a salt-resistant Saccharomyces cerevisiae (S. cerevisiae) mutant generated by evolutionary engineering. NaCl-resistant S. cerevisiae strains were obtained by ethyl methanesulfonate (EMS) mutagenesis followed by successive batch cultivations in the presence of gradually increasing NaCl concentrations, up to 8.5% w/v of NaCl (1.45 M). The most probable number (MPN) method, high-performance liquid chromatography (HPLC), and glucose oxidase/peroxidase method were used for physiological analysis, while Agilent yeast DNA microarray systems were used for transcriptome analysis. NaCl-resistant mutant strain T8 was highly cross-resistant to LiCl and highly sensitive to AlCl3. In the absence of NaCl stress, T8 strain had significantly higher trehalose and glycogen levels compared to the reference strain. Global transcriptome analysis by means of DNA microarrays showed that the genes related to stress response, carbohydrate transport, glycogen and trehalose biosynthesis, as well as biofilm formation, were upregulated. According to gene set enrichment analysis, 548 genes were upregulated and 22 downregulated in T8 strain, compared to the reference strain. Among the 548 upregulated genes, the highest upregulation was observed for the FLO11 (MUC1) gene (92-fold that of the reference strain). Overall, evolutionary engineering by chemical mutagenesis and increasing NaCl concentrations is a promising approach in developing industrial strains for biotechnological applications.
Keywords: Evolutionary engineering, transcriptome analysis, NaCl-resistance, Saccharomyces cerevisiae, stress resistance, industrial strains
INTRODUCTION
Maintaining alkali cation homeostasis in cells is crucial for many organisms, including yeasts. Yeast cells are exposed to stress in different industrial fermentation processes, such as baking, brewing, and winemaking. Improvement of salt tolerance in yeast would be beneficial for many industrial processes, e.g. the production of yeast biomass and baking [1]. Moreover, understanding molecular mechanisms of salt-tolerant yeasts can be useful for engineering salt-tolerant fungi and crop plants, since many components of their stress response systems are similar to those in yeasts [2]. Salt stress induces ion toxicity and osmotic stress in yeast cells [3], and has multiple effects on other cellular processes. High-salinity stress leads to high osmotic pressure that increases the flow of water out of the cell, resulting in lower internal pressure. Furthermore, high-salinity stress disrupts the membrane potential, which in turn affects the activity of membrane transporters and stability of ions and pH in the cell, leading to the generation of reactive oxygen species and misfolding of proteins [4].
The yeast Saccharomyces cerevisiae (S. cerevisiae) has relatively high concentrations of K+ in the cytoplasm and low concentrations of Na+. K+ is involved in many cell functions and is the most abundant cation in S. cerevisiae, even though K+ concentration is low in most of the natural habitats of yeast. In contrast, Na+ is abundant in many natural habitats; however, its accumulation within cells has a toxic effect. The balance between K+ and Na+ is regulated by the coordinated function of different transporters involved in influx and efflux of cations [5]. Yeast cells use several mechanisms for transferring ions through the cell membrane, including passive transport through ion channels, active transport by carrier proteins, proton countertransport, and transport mediated by adenosine 5’-triphosphatases (ATPases) [6]. S. cerevisiae has three different sodium transport systems: Nhx1p, Ena1-4p, and Nha1p. Nhx1p is an endosomal/prevacuolar Na+/H+ antiporter/exchanger, encoded by the NHX1 gene, and it increases the salt tolerance by moving Na+ and K+ to intracellular compartments, such as secretory organelles. Nha1p antiporter, localized in the plasma membrane, is encoded by the NHA1 gene and has substrate specificity for at least four cations, i.e., K+, Li+, Na+, and Rb+. Ena1-4p belongs to P-type Ena ATPases, it is located in the plasma membrane and also promotes the efflux of Na+, Li+, and K+ at high pH [7,8]. Moreover, the homeostasis of Na+, Li+, and K+ is maintained by all the three sodium transporters [9].
Yeast cells subjected to salt stress show dehydration, physiological and biochemical changes, and altered regulation of gene expression [10]. The cells develop tolerance to Na+ stress through ion homeostasis achieved by ion transport and detoxification mechanisms and through osmotic adjustment achieved by accumulating solutes inside the cell [2,11].
In this study, we performed a physiological and transcriptomic analysis of a salt-resistant S. cerevisiae mutant strain generated by evolutionary engineering. Evolutionary engineering is an inverse metabolic engineering approach for creating or improving the microbial phenotypes of interest [12]. Strains with specific characteristics and/or complex phenotypes can be obtained by evolutionary engineering, which is followed by high-throughput characterization and genome-wide microarray analysis. Evolutionary engineering strategies have been successfully applied in biofuel research [13] or for studying molecular mechanisms underlying stress resistance in different organisms [14-17].
We obtained NaCl-resistant S. cerevisiae strains by ethyl methanesulfonate (EMS) mutagenesis followed by successive batch cultivations in the presence of gradually increasing NaCl concentrations [18,19]. Yeast resistance to NaCl stress, at the physiological and transcriptomic level, was investigated in the mutant strain with the highest resistance to salt stress and compared to the reference strain.
MATERIALS AND METHODS
Yeast strain, culture reagents and conditions
S. cerevisiae strain CEN.PK113-7D (MATa, MAL2-8c, and SUC2) was used in this study [20]. Yeast extract peptone dextrose (1% w/v peptone, 1% w/v yeast extract, and 2% w/v glucose) and yeast minimal medium [YMM] (2% w/v glucose, 0.67% w/v yeast nitrogen base without amino acids [Difco, USA]) were used for cultivations. Yeast cultures were grown at 30°C and the growth was monitored with optical density measurements at 600 nm (OD600) using a Shimadzu UV-1700 spectrophotometer (Japan). The culture stocks were prepared in YMM containing 30% (v/v) glycerol and kept at −80°C.
Evolutionary engineering strategy for selecting NaCl-resistant S. cerevisiae mutant strains
S. cerevisiae CEN.PK 113-7D was used as the reference strain and subjected to EMS mutagenesis to obtain a genetically diverse population for selection. Successive batch cultures in the presence of gradually increasing NaCl concentrations were used to obtain NaCl-stress resistant S. cerevisiae mutants. To determine the initial NaCl concentration, both the reference strain and EMS-induced mutant population were screened at NaCl concentration in the range 0.17-1.7 M in YMM, during batch growth for 48 hours. The selection of cultures was started at 1% w/v of NaCl and increased up to 8.5% w/v (1.45 M). The isolation of mutant colonies in the selected NaCl-resistant population was performed on solid YMM with appropriate culture dilutions, followed by random isolation of mutant colonies after 48 hours of incubation at 30°C, as described previously [18].
Estimation of stress resistance
Yeast resistance to NaCl and other stress conditions was determined using spot test and most probable number (MPN) method [21]. For spot analysis, the cells were cultured in 10-ml liquid YMM and placed in 50-ml culture tubes at 30°C, until the logarithmic phase of growth was reached. Equal numbers of yeast cells corresponding to 4 OD600 units in 1-ml volume were harvested by centrifugation at 10,000 g for 5 minutes, and the supernatant was removed. The pellets were resuspended in 100 µl water and diluted serially from 10−1 to 10−4 in 96-well plates. Next, 2 µl of the diluted suspensions were placed on solid YMM plates (control) and YMM plates containing 0.7 M NaCl (Carlo Erba, Italy), 5 mM LiCl (Sigma, USA), 1 M KCl (Merck, Germany), 3 mM CoCl2 (Fluka, USA), 20 mM MnCl2 (Sigma, USA), 10 mM ZnCl2 (Sigma ALDRICH, USA), 0.15 mM CuCl2 (Sigma ALDRICH, USA), 1 mM H2O2 (Merck, Germany), 8% (v/v) ethanol (J.T. Baker, Netherlands), 2 mM CrCl3 (Acros Organics, USA) and 7.5 M AlCl3 (Merck, Germany), and incubated at 30°C for 48 hours.
For MPN analysis (Russek and Colwell, 1983), viable cell counts were made by serial dilutions in 96-well plates. Each well contained 180 µl YMM and 20 µl of cells. Dilutions were made in the range of 10−1 to 10−8 for five biological repeats. The published MPN tables (http://www.jlindquist.net/generalmicro/102dil3.html) were used to estimate the MPN of the cells, based on their growth ability at higher dilutions. The survival rate was assessed by dividing the number of viable cells in the presence of NaCl by the number of cells in the absence of NaCl. The S. cerevisiae reference strain and NaCl-resistant mutant were screened in different stress conditions, including: 0.7 M NaCl, 5 mM LiCl, 1.5 M KCl, 4 mM CoCl2, 15 mM MnCl2, 10 mM ZnCl2, 0.1 mM CuCl2, 0.8 mM H2O2, 6% (v/v) ethanol, 2 mM CrCl3, and 10 mM AlCl3.
High-performance liquid chromatography (HPLC) analysis of glucose, ethanol, acetate and glycerol
The reference and NaCl-resistant mutant strain were inoculated into 10 ml of YMM in 50-ml culture tubes to an OD600 of 0.05. After overnight incubation at 30°C, OD600 measurements were made using a spectrophotometer, and the cultures were inoculated into 100 ml fresh YMM in 500-ml flasks to an initial OD600 of 0.25. One control flask with YMM and two other flasks, one supplemented with 0.5 M NaCl in YMM and the other with 5 mM LiCl in YMM, were incubated at 30°C for 24 hours.
The experiments were performed in triplicate. OD600 values were measured at 1.5-hour intervals. From all cultures, 1 ml of samples was placed into microfuge tubes with known weight (for cell dry weight experiment) at 1.5-hour time intervals. After centrifugation (10,000 g for 5 minutes), the supernatants were filtered using a 0.2-µm pore filter (TPP Techno Plastic Products, Switzerland) and analyzed by HPLC (Shimadzu, Japan). Metabolite analysis by HPLC was performed using the Aminex© HPX-87H (Bio-Rad, USA) column eluted with H2SO4 (5 mM) at a flow rate of 0.6 ml/minute at 60°C, as described previously [16].
Enzymatic determination of glycogen and trehalose content
Glycogen and trehalose content of cultures were determined using an enzymatic method, as described by Parrou and François [22]. The glucose amount released from trehalose and glycogen was determined using the glucose oxidase/peroxidase method [23]. The glycogen and trehalose contents were quantitatively determined in the presence and absence of Na+ and Li+, in triplicate. Briefly, the cell pellet (obtained from 25 OD600 units of culture) was resuspended in 250 µl 0.25 M Na2CO3 in a screw top microcentrifuge tube (Corning, USA) and heated at 95°C for 4 hours. Afterward, 150 µl of 1 M acetic acid and subsequently 600 µl of 0.2 M sodium acetate buffer (pH 5.2) were added to the cell suspension. Half of this mixture was incubated overnight at 57°C in the presence of 100 mg of amyloglucosidase (Roche, Switzerland) in a hybridization chamber. The second half of the mixture (500 µl) was incubated overnight at 37°C in the presence of trehalase (Sigma, USA). The suspensions were centrifuged for 2 minutes at 5000 g. Then, 200 µl of glucose oxidase/peroxidase reagent (Sigma, USA) and 20 µl of supernatant was added to the wells of the 96-well plate. Following 30-minute incubation at 37°C, the absorbance of the samples was measured at 420/490 nm using a Benchmark Plus microplate reader (Bio-Rad, USA).
Whole genome transcriptome analysis of NaCl-resistant mutant and reference strain
The reference and NaCl-resistant mutant strains were grown in 500-ml flasks containing 100 ml YMM, at 30°C, until their OD600 values reached ~1 (5 × 107 cells/ml). Total RNA was extracted from the cultures using RNeasy Mini Kit (Qiagen, USA) according to the protocol described by the manufacturer, and stored at −80°C. The concentrations of extracted RNA were analyzed with a ultraviolet-visible (UV/VIS) spectrophotometer (NanoDrop 2000), and the quality of the extracted RNA was assessed using RNA 6000 Nano Kit (Agilent, USA) and BioAnalyzer 2100 (Agilent, USA). RNA samples with RNA integrity number (RIN) >8 were used in the transcriptome analysis. DNA microarray analysis was performed for the reference and mutant strain using Agilent DNA microarray systems (USA), in triplicate. One-color RNA spike-in kit (Agilent, USA) was used to provide internal controls with known concentrations. The total RNA samples were then mixed with diluted RNA spike-in controls and labeled with cyanine 3 (Cy3) using the Low-Input QuickAmp Labeling Kit (Agilent, USA). The Cy3-labelled samples were purified using the Absolutely RNA Nanoprep Kit (Agilent, USA), applied to the microarray slides (RNA 6000 Nano LabChip® Kit, Agilent, USA), and incubated. The microarray slides were transferred to a microarray hybridization chamber (Agilent, USA) for hybridization process at 65°C for 17 hours. The slides were then washed and scanned, according to the manufacturer’s instructions. Transcriptome data analysis was performed using GeneSpring GX software, version 12.5 (Agilent, USA). Reliability of the gene expression data was assessed by Student’s t-test and Benjamini and Hochberg [24] false discovery rate (FDR) correction. Significant differences in the gene expression were determined for the results with corrected p values that were <0.05. Cluster analysis was performed for genes exhibiting at least 2-fold change (increase and decrease) in the expression and having a p ≤ 0.01. The genes whose expression changed more than 2-fold were classified into clusters and functional categories using FunSpec online software [25] and FunCat database (http://mips.gsf.de/funcatDB/) [26].
This work is fully MIAME (Minimum Information About a Microarray Experiment)-compliant and has been deposited at GEO with the accession number GSE 61903 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE61903).
RESULTS
Selection of NaCl-resistant population by evolutionary engineering strategy
Initially, the EMS-induced mutant population and the reference strain were exposed to concentrations of NaCl in the range 0.1-1.7 M to determine the starting concentration of NaCl for the selection of salt-resistant strains. Based on the survival rates of the yeasts, the initial NaCl concentration was selected as 1% w/v (data not shown). The EMS-induced population was exposed to gradually increasing concentrations of NaCl in successive batch cultures. NaCl was present in the culture medium during the entire cultivation period, as a continuously applied stress factor. Forty passages of cells (about 160 generations) were obtained up to the final NaCl concentration of 8.5% w/v, corresponding to 1.45 M NaCl, as described previously [18].
A total of 23 individual mutant colonies were then randomly isolated on solid YMM plates, from the mutant population that was resistant to 8.5% w/v of NaCl. The 23 mutant colonies were initially tested on YMM plates containing 8% w/v of NaCl, and 10/23 colonies, named T4, T8, T10, T11, T12, T13, T15, T19, T20, and T23, had a higher resistance to NaCl (data not shown). The levels of resistance to NaCl in the 10 colonies were determined quantitatively using the MPN method, in the presence of 8% w/v of NaCl. The MPN results revealed that T8 mutant strain had the highest NaCl resistance, approximately 700-fold that of the reference strain (Figure 1). T8 was also genetically stable, based on 10 successive batch cultivations under non-selective culture conditions and according to the NaCl-resistance analysis after each cultivation (data not shown).
FIGURE 1.
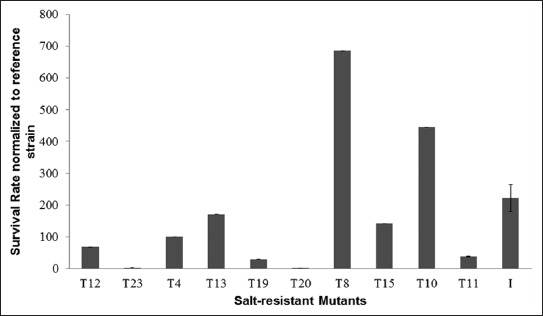
Survival rates of salt-resistant yeast mutant strains as determined by 5-tube most probable number method at the 72nd hour of incubation in yeast minimal medium containing 8% w/v of NaCl. “I” indicates an arithmetic average of the survival rates in mutant strains. Among salt resistant mutants, T8 yeast mutant strain had the highest survival rate.
Cross-resistance analysis of NaCl-resistant mutant strain
To test whether T8 mutant strain was cross-resistant to other stress factors, it was grown on solid media in different stress conditions. The results of spot assay revealed that T8 was also highly resistant to 5 mM LiCl, but highly sensitive to 7.5 mM AlCl3, compared to the reference strain. Moreover, T8 was slightly more resistant to 1 mM H2O2 and 10 mM ZnCl2, compared to the reference strain (Figure 2). The cross-resistance levels of T8 and the reference strain were then quantified by MPN method and these results confirmed data obtained with the spot assay as follows (Figure 3): T8 was hyper-resistant to 5 mM LiCl, with a survival rate of about 24,000-fold of the reference strain survival rate. It was also cross-resistant to 10 mM ZnCl2, 0.8 mM H2O2, 4 mM CoCl2, 1 M KCl, and 6% v/v ethanol. Furthermore, T8 was hypersensitive to 10 mM AlCl3, and slightly sensitive to 15 mM MnCl2 and 2 mM CrCl3. The MPN and spot test results for 1 M KCl and 0.15 mM CuCl2 were identical for the reference strain and T8. Overall, the MPN results were in accordance with the spot test results (Figure 3).
FIGURE 2.
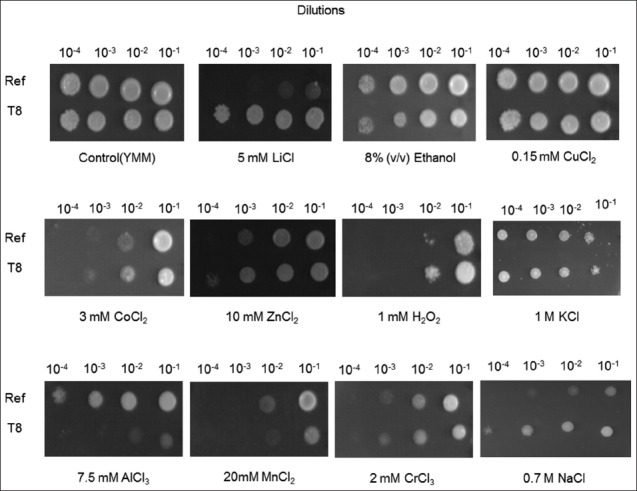
Analysis of cross-resistance in T8 mutant yeast strain at 48 hours of incubation on solid yeast minimal medium plates with stress factors diluted at different concentrations, as indicated in the figure. Ref represents the reference strain. T8 mutant yeast strain showed high resistance to NaCl and LiCl and high sensitivity to AlCl3.
FIGURE 3.
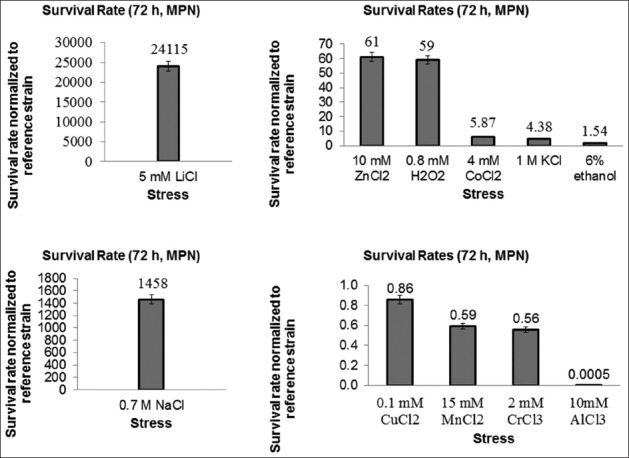
Survival rates of T8 mutant yeast strain normalized to those of the reference strain under stress conditions as follows: 5 mM LiCl, 0.7 M NaCl, 10 mM ZnCl2, 0.8 mM H2O2, 4 mM CoCl2, 1 M KCl, 6% (v/v) ethanol, 0.1 mM CuCl2, 15 mM MnCl2, 2 mM CrCl3, and 10 mM AlCl3. The survival rates were determined using the most probable number (MPN) method, at 72 hours of incubation in yeast minimal medium containing the stress factors. T8 was hyper-resistant to 5 mM LiCl and also cross-resistant to 10 mM ZnCl2, 0.8 mM H2O2, 4 mM CoCl2, 1 M KCl, and 6% v/v ethanol. The MPN results were in accordance with the spot test results.
Metabolite profiles during batch cultivation
The cell growth of T8 in the presence of NaCl and LiCl was significantly improved compared to the reference strain [19]. The glucose consumption, ethanol, glycerol and acetate production were monitored and shown in Figure 4.
FIGURE 4.
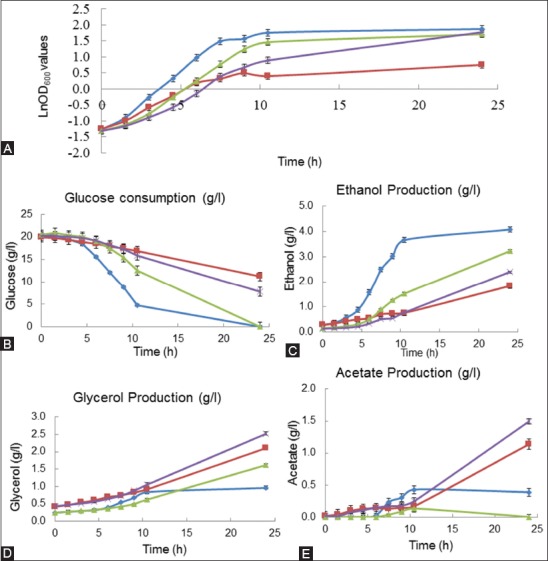
Growth behavior and metabolite profiles of the reference strain and T8 mutant yeast strain in yeast minimal medium, in the absence and presence of 0.5 M NaCl. (A) Growth behavior was monitored using optical density (OD) measurements at 600 nm, (B) glucose consumption (g/L), (C) ethanol production (g/L), (D) glycerol production (g/L), and (E) acetate production (g/L). The metabolite analyses were performed using high-performance liquid chromatography. “Filled diamond” represents reference strain, “filled square” represents reference strain in the presence of 0.5 M NaCl, “filled triangle” represents T8, and “filled circle” represents T8 in the presence of 0.5 M NaCl. T8 mutant yeast strain showed high growth rate in yeast minimal medium in the absence and presence of NaCl.
The growth analysis of T8 and the reference strain revealed that under 0.5 M NaCl the maximum specific growth rate (µmax) of the reference strain significantly decreased from 0.41 to 0.18 h−1. However, the µmax value of T8 decreased from 0.36 h−1 under control conditions to 0.28 h−1 under 0.5 M NaCl stress.
Interestingly, a significantly higher difference in the µmax values was observed between the reference strain and T8 under 5 mM LiCl stress. T8 had a µmax value of 0.38 h−1, whereas the reference strain had 0.09 h−1, indicating that T8 was not inhibited by 5 mM LiCl, despite the stronger inhibition of the reference strain by the LiCl compared to NaCl stress conditions. The glucose consumption of T8 was slightly higher compared to the reference strain, in the presence of 0.5 M NaCl (Figure 4). The presence of 0.5 M NaCl significantly decreased the ethanol production in both strains. Moreover, under the control conditions, T8 had lower ethanol production compared with the reference strain (Figure 4). In general, glycerol production was higher in T8 compared to the reference strain, in the presence and absence of NaCl stress; the highest glycerol levels in T8 were observed under NaCl stress (Figure 4). The presence of 0.5 M NaCl stress increased acetate production in both T8 and the reference strain, with higher levels of acetate produced by T8 compared to the reference strain. In the absence of NaCl, the reference strain had higher acetate production levels than T8 (Figure 4). The acetate production in T8 was the same under LiCl stress and control conditions. T8 reached the maximum acetate levels at 10 hours of growth, when glucose was depleted. After that, the acetate levels started to decrease in T8, both in the presence and absence of LiCl (Figure 5).
FIGURE 5.
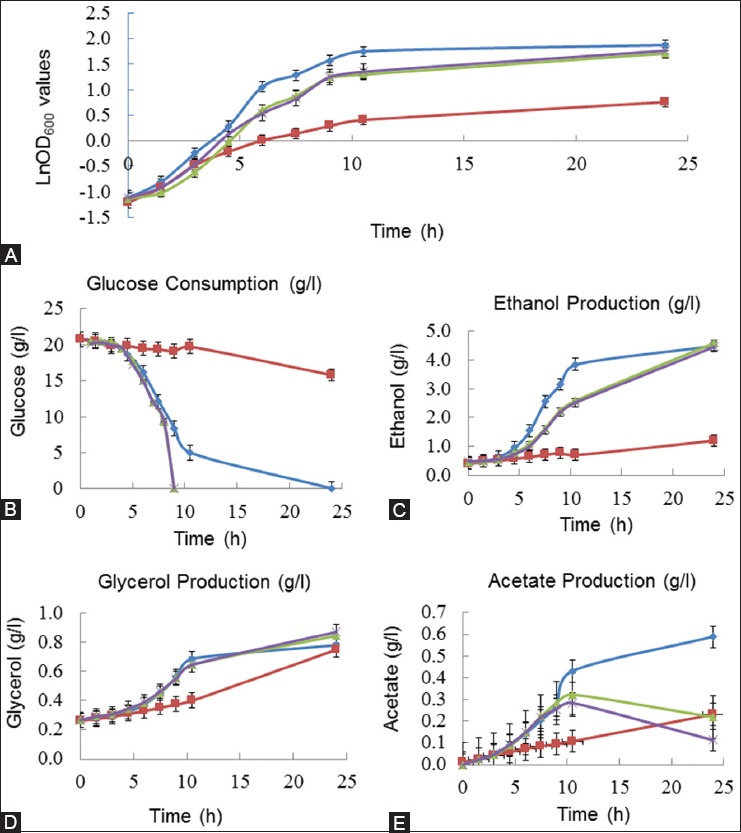
Growth behavior and metabolite profiles of the reference yeast and mutant T8 strain in yeast minimal medium, in the absence and presence of 5 mM LiCl. (A) Growth behavior was monitored using optical density measurements at 600 nm, (B) glucose consumption (g/L), (C) ethanol production (g/L), (D) glycerol production (g/L), and (E) acetate production (g/L). Metabolite analyses were performed using high-performance liquid chromatography. “Filled diamond” represents the reference strain, “filled square” represents the reference strain in the presence of 5 mM LiCl, “filled triangle” represents T8, and “filled circle” represents T8 in the presence of 5 mM LiCl. T8 mutant yeast strain showed an increase in growth rate with LiCl despite the stronger inhibition of the reference strain by LiCl.
Production of storage carbohydrates
Storage carbohydrate (trehalose and glycogen) levels were also determined in T8 and the reference strain, grown in the presence and absence of 0.5 M NaCl. In the absence of stress conditions, T8 had significantly higher trehalose levels (0.23 mg/mg) compared to the reference strain (0.01 mg/mg). The glycogen levels in T8 were also higher (0.09 mg/mg) under control conditions in relation to that of the reference strain (0.02 mg/mg). The presence of both NaCl and LiCl stress significantly increased glycogen and trehalose levels in the reference strain, and this was not observed in T8.
Whole genome transcriptome analysis
We performed the whole genome transcriptome analysis of the reference strain and the salt-resistant mutant T8. The statistical analyses showed that 548 genes were upregulated, and 22 genes were downregulated in T8, with 2-fold expression change (log2[fold change] ≥2), t-test p < 0.05. Those genes were classified into clusters and functional categories by FunSpec online software [25] and FUNCAT database [26]. According to this classification, the genes related to carbohydrate transport, glycogen and trehalose biosynthesis, glycolysis, phosphorylation, and biofilm formation were upregulated, whereas genes related to protein synthesis and transcription were downregulated in T8 (Table 1 and 2). The highest upregulation (92-fold that of the reference strain) in T8 was observed for the FLO11 (MUC1) gene, which encodes glycosylphosphatidylinositol (GPI)-anchored cell surface glycoprotein (flocculin, Flo11p). The Flo11p domain structure is similar to that of adhesins in pathogenic fungi. Flo11p is localized to the cell surface, and when cell–cell adhesion is induced, Flo11p is necessary for invasive growth, biofilm formation, and pseudohyphal development [27,28]. Hexokinase isoenzyme 1 (HXK1) was also upregulated in T8 (approximately 90-fold). Similarly, other highly upregulated genes (approximately 30-fold) were related to hexose transport. Furthermore, the GPD1 gene (glycerol synthesis) and TSL1, TPS1, TPS3, TPS2, and NTH1 (all related to trehalose synthesis) along with acid trehalose gene (ATH1) were upregulated in T8 by more than 2-fold.
TABLE 1.
Upregulated genes in NaCl-resistant T8 yeast mutant strain (5-fold and higher compared to the reference strain)
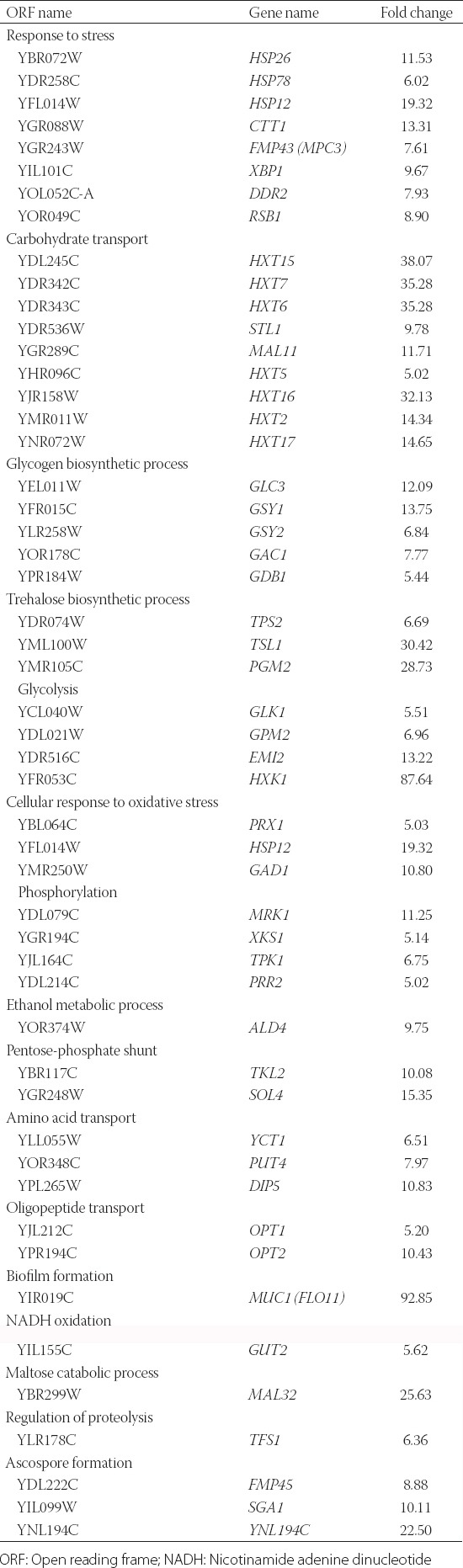
TABLE 2.
Downregulated genes in NaCl-resistant T8 yeast mutant strain (2-fold and higher compared to the reference strain)
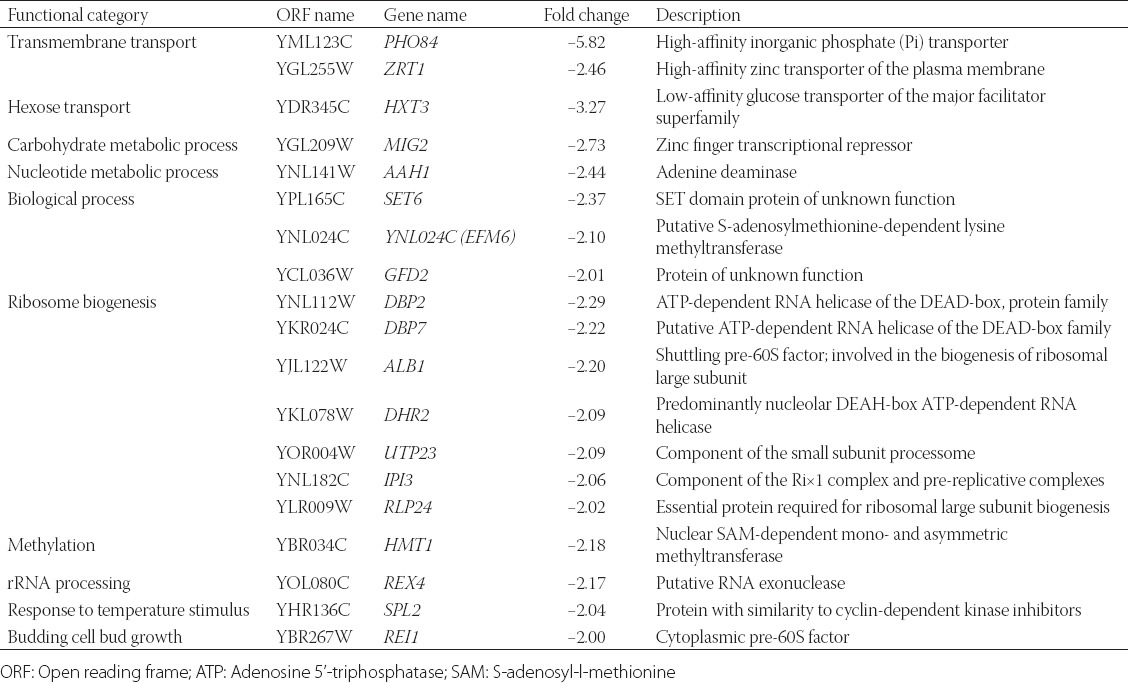
Different signaling pathways may be involved in the regulation of the response to osmotic stress, including the ENA1 and GPD1 genes. The vacuolar-type H+-ATPase (V-ATPase) subunit genes (e.g., VMA6, VPH1, VMA7, VMA5, and TFP1 [VMA1]), involved in sequestration of sodium into the vacuole, were not upregulated in T8. The genes involved in the synthesis and degradation of glycogen, such as UGP1, GAC1, GPH1, GSY1, GLC3, and GLG1, were upregulated in T8 by at least 4-fold under NaCl stress.
DISCUSSION
Evolutionary engineering is a promising and powerful strategy for the development of yeast strains. Based on a previous study [18], we obtained NaCl-resistant S. cerevisiae strains by EMS mutagenesis followed by successive batch cultivations in the presence of gradually increasing NaCl concentrations, up to 8.5% w/v of NaCl (1.45 M). The NaCl-resistant strain T8 could tolerate the highest, 1.45 M, concentration of NaCl; its resistance to NaCl was 700-fold that of the reference strain [18]. Yeast strains have been improved based on a long-term adaptation of yeast to environmental or metabolic constraints [2,16,17,29-33]. Among those studies, Gerstein et al. [28] and Dhar et al. [2] also included experimental evolution experiments with NaCl. Gerstein et al. [28] showed convergent evolution toward diploidy, in initially haploid and tetraploid lines of S. cerevisiae, under both unstressful and stressful conditions (0.6 M NaCl). Dhar et al. [2] evolved three replicate lines of S. cerevisiae BY4741 strain under continuous NaCl stress (0.5 M), and showed that the adaptation of yeast to salt stress is associated with genome size increase and moderate changes in the expression of several genes [2].
Our analysis of cross-resistance revealed that T8 mutant strain was highly cross-resistant to 5 mM LiCl and moderately cross-resistant to ZnCl2, H2O2, CoCl2, KCl, and EtOH stress. Moreover, T8 was highly sensitive to AlCl3. The sensitivity of T8 to aluminum is an example of trade-off in evolutionary engineering, where gaining a new trait may cause the loss or weakening of another trait, as explained previously [32]. In a previous study, S. cerevisiae FY73 strain which was transformed with the YEpGALR1 (+ALR1) or YEpGALR2 (+ALR2) overexpression construct had increased tolerance to Al+3 and Ga+3 but also showed increased sensitivity to Zn+2, Mn+2, Ni+2, Cu+2, Ca+2, and La+3 [33]. This is in agreement with our results, as T8 was moderately cross-resistant to Co+2, Ni+2, Zn+2 and sensitive to Mn+2 and Al+3. It was also shown that NaCl improves zinc tolerance in S. cerevisiae and Zygosaccharomyces rouxii [34], which correlates with our results, as T8 was cross-resistant to ZnCl2.
The homeostasis of Li+ and Na+ in yeast is maintained by the same multiple transport pathways [35]. While high concentrations of Li+ cause cation toxicity and high K+ concentrations cause hyperosmotic stress, high NaCl stress leads to both cation toxicity and hyperosmotic stress. Na+ is less toxic than Li+, however more toxic than K+ [36,37]. The high resistance of T8 to Na+ and Li+ in our experiments, suggests that T8 had resistance to cation toxicity rather than to osmotic stress.
The CEN.PK family is more hypersensitive to Na+ and Li+ compared to other S. cerevisiae strains [38,39]. Daran-Lapujade et al. [40] indicated that the PMR2 locus in CEN.PK113-7D harbours a single gene (ENA6), which encodes plasma membrane sodium ATPase. The ENA6 is similar to ENA1, 2 and 5 in other yeast strains. The authors concluded that the hypersensitivity of CEN.PK113-7D to Na+ and Li+ is associated with the role of Ena6 as the major exporter of those cations. When the ENA6 was overexpressed, the tolerance of CEN.PK113-7D to Na+ and Li+ was restored. Moreover, while CEN.PK113-7D was sensitive to 10 mM LiCl and 500 mM NaCl, our T8 mutant strain was resistant to those stress conditions.
Glycogen and trehalose are two important storage metabolites in S. cerevisiae and their production changes in response to a number of environmental stress conditions [11]. S. cerevisiae responds to salt stress by accumulating the disaccharide trehalose. The expression of the enzymes required for trehalose synthesis, from trehalose-6-phosphate synthase to trehalase, is induced under salt stress, resulting in increased production of trehalose. For example, the TPS1 gene encoding trehalose-6-phosphate synthase was highly upregulated in yeast under salt stress [41]. On the contrary, our transcriptome analysis did not show the upregulation of TPS1 in T8, but other genes involved in trehalose synthesis, such as TPS2, TSL1, PGM2, UGP1, GLK1 and ATH1, were upregulated in T8 under salt stress. Moreover, the glycogen and trehalose content was significantly higher in T8 compared to the reference strain, even in the absence of NaCl stress. Our results also revealed that Li+ triggered higher accumulation of glycogen and trehalose compared to Na+, which may be due to the higher toxicity of Li+. The accumulation of trehalose was higher than the accumulation of glycogen in T8, both in the presence and absence of Na+ and Li+. In general, high tolerance to Li+ may be due to effective efflux of Li+, and the Ena ATPase is the most powerful system for the export of alkali metal cations [42]. In our study, the ENA6 was upregulated in T8 by approximately 8-fold.
High-osmolarity glycerol (HOG) signaling pathway is crucial in global stress response. However, the HOG1 gene was not upregulated in our T8 mutant strain. The HOG signaling pathway is important in the transcriptional response of most salt-responsive genes but other signaling pathways may also be involved [43].
Dhar et al. [2] demonstrated changes in the expression of a number of well-known stress-response genes, such as the CTT1, MSN4, and HLR1. They showed that the basal expression of CTT1 in S. cerevisiae BY4741 mutant strain increased by 1.6-fold under NaCl stress, compared to the reference strain. The authors indicated that the adaptation of S. cerevisiae to NaCl stress was associated with changes in the expression of several genes [2]. Our transcriptome analysis of NaCl-resistant mutant T8 also revealed significant upregulation of CTT1, approximately 13-fold that of the reference strain, as well as of several other genes related to the stress response. However, no differences were observed between T8 and the reference strain in the expression levels of MSN4 and HLR1. Hyperosmotic stress has been shown to induce an oxidative stress response. Namely, the expression of the cytosolic catalase T gene CTT1 is known to increase along with the expression of multicopy suppressor of SNF1 mutation transcription factors Msn2, Msn4 and Msn1 in S. cerevisiae after the exposure to osmotic shock. The MSN2 and MSN4 genes are activated in various stress conditions [36,44]. In response to osmotic stress, Msn2 and Msn4 enter the nucleus and bind to the CTT1 promoter. Through the interaction with Msn2 and Msn4, Hog1 binds to DNA and activates the expression of CTT1 [45]. Consistent with the previous statements, we demonstrated a higher CTT1 expression in our NaCl-resistant mutant strain T8, compared to the reference strain. The highest upregulation was observed for the FLO11 (MUC1) gene (92-fold that of the reference strain). The FLO11 gene encodes flocculin which is responsible for the attachment of yeast cell to the surface or another cell, by binding to an amino acid or glucose molecule present on the cell surface [46]. Many of the HXT genes (i.e. HXT2, HXT6, HXT7, HXT15, HXT16, and HXT17), which are related to hexose transport, were highly upregulated in our T8 mutant strain; in addition, the HXK1 gene was the second highest upregulated gene in T8 (approximately 88-fold that of the reference strain). Specific genes that control the growth rate of S. cerevisiae help the organism to adapt to diverse environmental conditions [47]. Our T8 mutant strain probably responded to carbon limitation and controlled its growth by the upregulation of HXK1, GLK1, HXT6, and HXT7 gene (Table 1). The ADR1, CAT8, USV1, and MTH1 were also upregulated in T8, possibly in response to carbon metabolism regulation. On the contrary, we observed downregulation of low-affinity glucose transporter genes HXT1 and HXT3 in T8. Finally, the upregulation of genes related to the biosynthesis and degradation of trehalose, in the environment with limited carbon, was associated with an improved growth rate in T8. The described results are compatible with those reported by Gutteridge et al. [48]. The TPK2 and VHS3 genes were upregulated in T8 by more than 2-fold, as well as the PPZ2 (an isoform of PPZ1), but there were no differences in the expression levels of TRK1, TRK2, and PPZ1 compared to our reference strain.
Overall, evolutionary engineering by chemical mutagenesis and increasing NaCl concentrations is a promising approach in developing industrial strains for biotechnological applications. Because of various stress conditions in industrial processes, increasing the performance of yeast strains is necessary. With this regard, it is important to understand the molecular mechanisms of salt stress in yeast, as well as its relationship with other stress factors. The productivity of biotechnological processes involving yeast may be improved using salt-tolerant mutant strains.
ACKNOWLEDGMENTS
This work was supported by Istanbul Technical University Research Funds (ITU-BAP graduate student project no: 37730). S. Hande Tekarslan Sahin was supported by The Scientific and Technological Research Council of Turkey – National Scholarship Programme for PhD students. The authors would like to thank professors Petek Cakar and Carola Hunte for their scientific support. The authors wish to thank H. Ibrahim Kisakesen, Mehmet Ali Tekin, and Nilay Ordek for technical help and Dr. Xenia Bogdanovic for editing the manuscript.
DECLARATION OF INTERESTS
The authors declare no conflict of interests.
REFERENCES
- 1.Attfield PV. Stress tolerance: the key to effective strains of industrial baker’s yeast. Nat Biotechnol. 1997;15(13):1351–7. doi: 10.1038/nbt1297-1351. https://doi.org/10.1038/nbt1297-1351. [DOI] [PubMed] [Google Scholar]
- 2.Dhar R, Sägesser R, Weikert C, Yuan J, Wagner A. Adaptation of Saccharomyces cerevisiae to saline stress through laboratory evolution. J Evol Biol. 2011;24(5):1135–53. doi: 10.1111/j.1420-9101.2011.02249.x. https://doi.org/10.1111/j.1420-9101.2011.02249.x. [DOI] [PubMed] [Google Scholar]
- 3.Posas F, Chambers JR, Heyman JA, Hoeffler JP, de Nadal E, Ariño J. The transcriptional response of yeast to saline stress. J Biol Chem. 2000;275(23):17249–55. doi: 10.1074/jbc.M910016199. https://doi.org/10.1074/jbc.M910016199. [DOI] [PubMed] [Google Scholar]
- 4.Melamed D, Pnueli L, Arava Y. Yeast translational response to high salinity: global analysis reveals regulation at multiple levels. RNA. 2008;14(7):1337–51. doi: 10.1261/rna.864908. https://doi.org/10.1261/rna.864908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bañuelos MA, Ruiz MC, Jiménez A, Souciet JL, Potier S, Ramos J. Role of the Nha1 antiporter in regulating K(+) influx in Saccharomyces cerevisiae . Yeast. 2002;19(1):9–15. doi: 10.1002/yea.799. https://doi.org/10.1002/yea.799. [DOI] [PubMed] [Google Scholar]
- 6.Serrano R, Rodriguez-Navarro A. Ion homeostasis during salt stress in plants. Curr Opin Cell Biol. 2001;13(4):399–404. doi: 10.1016/s0955-0674(00)00227-1. https://doi.org/10.1016/S0955-0674(00)00227-1. [DOI] [PubMed] [Google Scholar]
- 7.Cyert MS, Philpott CC. Regulation of cation balance in Saccharomyces cerevisiae . Genetics. 2013;193(3):677–713. doi: 10.1534/genetics.112.147207. https://doi.org/10.1534/genetics.112.147207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hunte C, Screpanti E, Venturi M, Rimon A, Padan E, Michel H. Structure of a Na+/H+antiporter and insights into mechanism of action and regulation by pH. Nature. 2005;435(7046):1197–202. doi: 10.1038/nature03692. https://doi.org/10.1038/nature03692. [DOI] [PubMed] [Google Scholar]
- 9.Kinclova-Zimmermannova O, Gaskova D, Sychrova H. The Na+K+/H+-antiporter Nha1 influences the plasma membrane potential of Saccharomyces cerevisiae . FEMS Yeast Res. 2006;6(5):792–800. doi: 10.1111/j.1567-1364.2006.00062.x. https://doi.org/10.1111/j.1567-1364.2006.00062.x. [DOI] [PubMed] [Google Scholar]
- 10.Mager WH, Siderius M. Novel insights into the osmotic stress response of yeast. FEMS Yeast Res. 2002;2(3):251–7. doi: 10.1016/S1567-1356(02)00116-2. https://doi.org/10.1016/S1567-1356(02)00116-2. [DOI] [PubMed] [Google Scholar]
- 11.François JM, Walther T, Parrou JL, Liu ZL. Microbial Stress Tolerance for Biofuels. Berlin, Heidelberg: Springer-Verlag; 2012. Genetics and regulation of glycogen and trehalose metabolism in Saccharomyces cerevisiae; pp. 29–55. https://doi.org/10.1007/978-3-642-21467-7_2. [Google Scholar]
- 12.Patnaik R. Engineering complex phenotypes in industrial strains. Biotechnol Prog. 2008;24(1):38–47. doi: 10.1021/bp0701214. https://doi.org/10.1021/bp0701214. [DOI] [PubMed] [Google Scholar]
- 13.Koppram R, Albers E, Olsson L. Evolutionary engineering strategies to enhance tolerance of xylose utilizing recombinant yeast to inhibitors derived from spruce biomass. Biotechnol Biofuels. 2012;5:1–32. doi: 10.1186/1754-6834-5-32. https://doi.org/10.1186/1754-6834-5-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cakar ZP, Seker UO, Tamerler C, Sonderegger M, Sauer U. Evolutionary engineering of multiple-stress resistant Saccharomyces cerevisiae. FEMS Yeast Res. 2005;5(6-7):569–78. doi: 10.1016/j.femsyr.2004.10.010. https://doi.org/10.1016/j.femsyr.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 15.Cakar ZP, Alkim C, Turanli B, Tokman N, Akman S, Sarikaya M, et al. Isolation of cobalt hyper-resistant mutants of Saccharomyces cerevisiae by in vivo evolutionary engineering approach. J Biotechnol. 2009;143(2):130–8. doi: 10.1016/j.jbiotec.2009.06.024. https://doi.org/10.1016/j.jbiotec.2009.06.024. [DOI] [PubMed] [Google Scholar]
- 16.Küçükgöze G, Alkım C, Yılmaz Ü, Kısakesen HI, Gündüz S, Akman S, et al. Evolutionary engineering and molecular characterization of nickel-resistant Saccharomyces cerevisiae . FEMS Yeast Res. 2013;13(8):731–46. doi: 10.1111/1567-1364.12073. https://doi.org/10.1111/1567-1364.12073. [DOI] [PubMed] [Google Scholar]
- 17.Alkim C, Benbadis L, Yilmaz U, Cakar ZP, François JM. Mechanisms other than activation of the iron regulon account for the hyper-resistance to cobalt of a Saccharomyces cerevisiae strain obtained by evolutionary engineering. Metallomics. 2013;5(8):1043–60. doi: 10.1039/c3mt00107e. https://doi.org/10.1039/c3mt00107e. [DOI] [PubMed] [Google Scholar]
- 18.Sezgin T. Evolutionary Engineering of Saccharomyces cerevisiae for Improved Industrial Properties. [PhD Thesis] Turkey: Istanbul Technical University; 2010. p. 124. [Google Scholar]
- 19.Tekarslan SH, Alkım C, Hunte C, Cakar ZP. Physiological and genetic analysis of cellular sodium and lithium response/resistance behavior using the yeast Saccharomyces cerevisiae as a model organism. Istanb J Pharm. 2015;45(2):165–79. [Google Scholar]
- 20.Entian KD, Kötter P. 25 yeast genetic strain and plasmid collections. Meth Microbiol. 2007;36:629–66. https://doi.org/10.1016/S0580-9517(06)36025-4. [Google Scholar]
- 21.Russek E, Colwell RR. Computation of most probable numbers. Appl Environ Microbiol. 1983;45(5):1646–50. doi: 10.1128/aem.45.5.1646-1650.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parrou JL, François J. A simplified procedure for a rapid and reliable assay of both glycogen and trehalose in whole yeast cells. Anal Biochem. 1997;248(1):186–8. doi: 10.1006/abio.1997.2138. https://doi.org/10.1006/abio.1997.2138. [DOI] [PubMed] [Google Scholar]
- 23.Cramp DG. New automated method for measuring glucose by glucose oxidase. J Clin Pathol. 1967;20(6):910–2. doi: 10.1136/jcp.20.6.910. https://doi.org/10.1136/jcp.20.6.910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57(1):289–300. [Google Scholar]
- 25.Robinson MD, Grigull J, Mohammad N, Hughes TR. FunSpec: a web-based cluster interpreter for yeast. BMC Bioinformatics. 2002;3:35. doi: 10.1186/1471-2105-3-35. https://doi.org/10.1186/1471-2105-3-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruepp A, Zollner A, Maier D, Albermann K, Hani J, Mokrejs M, et al. The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Res. 2004;32(18):5539–45. doi: 10.1093/nar/gkh894. https://doi.org/10.1093/nar/gkh894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Braus GH, Grundmann O, Brückner S, Mösch HU. Amino acid starvation and Gcn4p regulate adhesive growth and FLO11 gene expression in Saccharomyces cerevisiae. Mol Biol Cell. 2003;14(10):4272–84. doi: 10.1091/mbc.E03-01-0042. https://doi.org/10.1091/mbc.E03-01-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gerstein AC, Chun HJ, Grant A, Otto SP. Genomic convergence toward diploidy in Saccharomyces cerevisiae. PLoS Genet. 2006;2(9):e145. doi: 10.1371/journal.pgen.0020145. https://doi.org/10.1371/journal.pgen.0020145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wisselink HW, Torikens MJ, Wu Q, Pronk JT, van Maris AJ. Novel evolutionary engineering approach for accelerated utilization of glucose, xylose, and arabinose mixtures by engineered Saccharomyces cerevisiae strains. Appl Environ Microbiol. 2009;75(4):907–14. doi: 10.1128/AEM.02268-08. https://doi.org/10.1128/AEM.02268-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stanley D, Bandara A, Fraser S, Chambers PJ, Stanley GA. The ethanol stress response and ethanol tolerance of Saccharomyces cerevisiae . J Appl Microbiol. 2010;109(1):13–24. doi: 10.1111/j.1365-2672.2009.04657.x. https://doi.org/10.1111/j.1365-2672.2009.04657.x. [DOI] [PubMed] [Google Scholar]
- 31.Kutyna DR, Varela C, Stanley GA, Borneman AR, Henschke PA, Chambers PJ. Adaptive evolution of Saccharomyces cerevisiae to generate strains with enhanced glycerol production. Appl Microbiol Biotechnol. 2012;93(3):1175–84. doi: 10.1007/s00253-011-3622-7. https://doi.org/10.1007/s00253-011-3622-7. [DOI] [PubMed] [Google Scholar]
- 32.Wenger JW, Piotrowski J, Nagarajan S, Chiotti K, Sherlock G, Rosenzweig F. Hunger artists: Yeast adapted to carbon limitation show trade-offs under carbon sufficiency. PLoS Genet. 2011;7:e1002202. doi: 10.1371/journal.pgen.1002202. https://doi.org/10.1371/journal.pgen.1002202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.MacDiarmid CW, Gardner RC. Overexpression of the Saccharomyces cerevisiae magnesium transport system confers resistance to aluminum ion. J Biol Chem. 1998;273(3):1727–32. doi: 10.1074/jbc.273.3.1727. https://doi.org/10.1074/jbc.273.3.1727. [DOI] [PubMed] [Google Scholar]
- 34.Li C, Xu Y, Jiang W, Dong X, Wang D, Liu B. Effect of NaCl on the heavy metal tolerance and bioaccumulation of Zygosaccharomyces rouxii and Saccharomyces cerevisiae . Bioresour Technol. 2013;143:46–52. doi: 10.1016/j.biortech.2013.05.114. https://doi.org/10.1016/j.biortech.2013.05.114. [DOI] [PubMed] [Google Scholar]
- 35.Masuda CA, Ramírez J, Peña A, Montero-Lomelí M. Regulation of monovalent ion homeostasis and pH by the Ser-Thr protein phosphatase SIT4 in Saccharomyces cerevisiae . J Biol Chem. 2000;275(40):30957–61. doi: 10.1074/jbc.M004869200. https://doi.org/10.1074/jbc.M004869200. [DOI] [PubMed] [Google Scholar]
- 36.Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, et al. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11(12):4241–57. doi: 10.1091/mbc.11.12.4241. https://doi.org/10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marešová L, Sychrová H. Genetic interactions among the Arl1 GTPase and intracellular Na+/H+antiporters in pH homeostasis and cation detoxification. FEMS Yeast Res. 2010;10(7):802–11. doi: 10.1111/j.1567-1364.2010.00661.x. https://doi.org/10.1111/j.1567-1364.2010.00661.x. [DOI] [PubMed] [Google Scholar]
- 38.Garciadeblas B, Rubio F, Quintero FJ, Bañuelos MA, Haro R, Rodríguez-Navarro A. Differential expression of two genes encoding isoforms of the ATPase involved in sodium efflux in Saccharomyces cerevisiae. Mol Gen Genet. 1993;236(2-3):363–8. doi: 10.1007/BF00277134. https://doi.org/10.1007/BF00277134. [DOI] [PubMed] [Google Scholar]
- 39.Wieland J, Nitsche AM, Strayle J, Steiner H, Rudolph HK. The PMR2 gene cluster encodes functionally distinct isoforms of a putative Na+pump in the yeast plasma membrane. EMBO J. 1995;14(16):3870–82. doi: 10.1002/j.1460-2075.1995.tb00059.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Daran-Lapujade P, Daran JM, Luttik MA, Almering MJ, Pronk JT, Kotter P. An atypical PMR2 locus is responsible for hypersensitivity to sodium and lithium cations in the laboratory strain Saccharomyces cerevisiae CEN.PK113-7D. FEMS Yeast Res. 2009;9(5):789–92. doi: 10.1111/j.1567-1364.2009.00530.x. https://doi.org/10.1111/j.1567-1364.2009.00530.x. [DOI] [PubMed] [Google Scholar]
- 41.Blomberg A. Metabolic surprises in Saccharomyces cerevisiae during adaptation to saline conditions: questions, some answers and a model. FEMS Microbiol Lett. 2000;182(1):1–8. doi: 10.1111/j.1574-6968.2000.tb08864.x. https://doi.org/10.1111/j.1574-6968.2000.tb08864.x. [DOI] [PubMed] [Google Scholar]
- 42.Bubnová M, Zemancíková J, Sychrová H. Osmotolerant yeast species differ in basic physiological parameters and in tolerance of non-osmotic stresses. Yeast. 2014;31(8):309–21. doi: 10.1002/yea.3024. https://doi.org/10.1002/yea.3024. [DOI] [PubMed] [Google Scholar]
- 43.Zara G, Zara S, Pinna C, Marceddu S, Budroni M. FLO11 gene length and transcriptional level affect biofilm-forming ability of wild flor strains of Saccharomyces cerevisiae. Microbiology. 2009;155(Pt 12):3838–46. doi: 10.1099/mic.0.028738-0. https://doi.org/10.1099/mic.0.028738-0. [DOI] [PubMed] [Google Scholar]
- 44.Kobayashi N, McEntee K. Evidence for a heat shock transcription factor-independent mechanism for heat shock induction of transcription in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1990;87(17):6550–4. doi: 10.1073/pnas.87.17.6550. https://doi.org/10.1073/pnas.87.17.6550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O’Rourke SM, Herskowitz I, O’Shea EK. Yeast go the whole HOG for the hyperosmotic response. Trends Genet. 2002;18(8):405–12. doi: 10.1016/s0168-9525(02)02723-3. https://doi.org/10.1016/S0168-9525(02)02723-3. [DOI] [PubMed] [Google Scholar]
- 46.Claro FB, Rijsbrack K, Soares EV. Flocculation onset in Saccharomyces cerevisiae: effect of ethanol, heat and osmotic stress. J Appl Microbiol. 2007;102(3):693–700. doi: 10.1111/j.1365-2672.2006.03130.x. https://doi.org/10.1111/j.1365-2672.2006.03130.x. [DOI] [PubMed] [Google Scholar]
- 47.Pir P, Gutteridge A, Wu J, Rash B, Kell DB, Zhang N, et al. The genetic control of growth rate: a systems biology study in yeast. BMC Syst Biol. 2012;6:4. doi: 10.1186/1752-0509-6-4. https://doi.org/10.1186/1752-0509-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gutteridge A, Pir P, Castrillo JI, Charles PD, Lilley KS, Oliver SG. Nutrient control of eukaryote cell growth: a systems biology study in yeast. BMC Biol. 2010;8:68. doi: 10.1186/1741-7007-8-68. https://doi.org/10.1186/1741-7007-8-68. [DOI] [PMC free article] [PubMed] [Google Scholar]


