Abstract
The proton-coupled oligopeptide transporter PHT1 (SLC15A4), which facilitates cross-membrane transport of histidine and small peptides from inside the endosomes or lysosomes to cytosol, plays an important role in intracellular peptides homeostasis and innate immune responses. However, it remains a challenge to elucidate functional properties of the PHT1 transporter because of its subcellular localization. The purpose of this study was to resort hPHT1 protein from the subcellular to outer cell membrane of MDCK cells stably transfected with human PHT1 mutants, and to characterize its functional activity in these cells. Using this model, the functional activity of hPHT1 was evaluated by cellular uptake studies with d3-l-histidine, GlySar, and the bacterial peptidoglycan products MDP and Tri-DAP. We found that the disruption of two dileucine motifs was indispensable for hPHT1 transporter being preferentially targeting to plasma membranes. hPHT1 showed high affinity for d3-l-histidine and low affinity for GlySar, with Km values of 16.3 ± 1.9 µM and 1.60 ± 0.30 mM, respectively. Moreover, the bacterial peptidoglycan components MDP and Tri-DAP were shown conclusively to be hPHT1 substrates. The uptake of MDP by hPHT1 was inhibited by di/tripeptides and peptide-like drugs, but not by glycine and acyclovir. The functional activity of hPHT1 was also pH-dependent, with an optimal cellular uptake in buffer pH 6.5. Taken together, we established a novel cell model to evaluate the function of hPHT1 in vitro, and confirmed that MDP and Tri-DAP were substrates of hPHT1. Our findings suggest that PHT1 may serve as a potential target for reducing the immune responses and for drug treatment of inflammatory diseases.
Keywords: hPHT1, mutation, l-histidine, bacterial peptidoglycans, functional characterization
Graphical abstract
1. INTRODUCTION
The proton-coupled oligopeptide transporters (POTs) utilize a proton gradient as the motive force for uphill transport of di/tripeptides and peptide-like compounds across membranes.1,2 The POT family consists of four mammalian members, namely PEPT1, PEPT2, PHT1, and PHT2, which are predicted to contain 12 transmembrane domains with N- and C-termini in the cytosol.2–4 PEPT1 (SLC15A1) and PEPT2 (SLC15A2) are expressed at the apical side of cell membranes and mediate the uptake of di/tripeptide substrates into intestinal and renal epithelia, respectively.5,6 PHT1 (SLC15A4) and PHT2 (SLC15A3) are significantly expressed in the brain and lymphatic system, and currently are considered as endosomal and lysosomal transporters with limited substrate specificity for histidine and certain oligopeptides.7,8 The substrate spectrum of PEPT1 and PEPT2 are reasonably understood, however, there is little information about the substrate spectrum of PHT1 and PHT2, even though they share many functional and molecular properties with the PEPT1 and PEPT2 transporters.
Several studies have suggested that PHT1 is associated with diabetes, inflammatory bowel disease, and systemic lupus erythematosus.9–12 Researchers have also reported that PHT1 plays a role in NOD and TLR ligands-triggered immune responses. For example, using genetically modified mice, cytokine production mediated by the NOD1 ligand Tri-DAP, was reduced in SLC15A4 deficient mice.13 Moreover, knockdown of PHT1 significantly decreased Tri-DAP induced NF-κB activation in HEK293T cells.10 Since NOD1 and NOD2 act as cytosolic pattern recognition receptors for bacterial peptidoglycans, they are essential for activation of the intracellular inflammatory signal pathway. NOD1 is able to detect many types of DAP, such as dipeptide iE-DAP, tripeptide Tri-DAP, and M-Tri-DAP, whereas NOD2 senses MDP.14–17 The stimulation of NOD1 and NOD2 activates NF-κB transcription and downstream signal transduction.18 Although NOD1 and NOD2 detect peptidoglycans in the cytosol, the mechanism by which such peptidoglycans enter the cytosol from endosomal and lysosomal compartments remains unclear. In mammalian cells, MDP and Tri-DAP can be transported into the cytosol by PEPT1 and PEPT2 located on extracellular membranes,19,20 and by PHT2 located on endosomal and/or lysosomal membranes.21 Similar to PHT2, it was found that PHT1 is located in the membranes of endosomes and lysosomes.7 These observations indicate that PHT1 might also mediate the transport of bacterial peptidoglycan motifs, such as MDP and Tri-DAP.
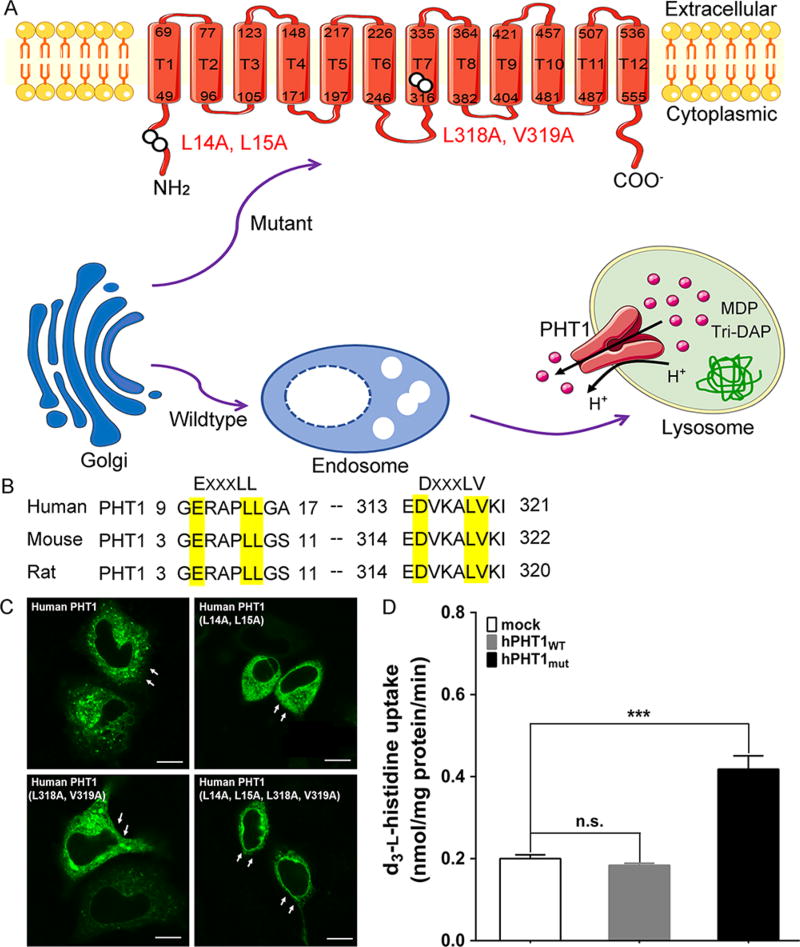
Using hPHT1 transiently transfected COS-7 cells, Bhardwaj et al. did not find the influx of GlySar in their uptake studies, possibly because of the subcellular location of PHT1.8,22 The subcellular location of endogenous hPHT1 makes it hard to characterize its transport kinetics and substrate specificity. To better understand the role of PHT1 in immune response, it is important to change the signals for sorting transmembrane proteins to endosomes and lysosomes. Currently, PHT2 has been successfully targeted to localize in plasma membranes by disruption of cytoplasmic dileucine motifs.21 The dileucine-based motif [DE]-XXXL-[LI] or DXXLL is responsible for transmembrane proteins being rapidly internalized and delivered to endosomes and lysosomes.7,23 Either of the two dileucine motifs substituted by alanine abrogates the lysosomal sorting.21,24 Since the amino acid sequence of hPHT1 shows good similarity to hPHT2 (50% amino acid identity) and also shares 80–90% identity to rat and mouse PHT1 proteins, there exists a molecular basis for the preferential targeting of PHT1 expression in plasma membranes.22,25,26 Still, no studies have constructed hPHT1 stably transfected mammalian cells and systematically evaluated its functional activity.
With this in mind, the objective of this study was to establish hPHT1 stably transfected MDCK cells and to evaluate the cellular uptake properties of histidine, GlySar, and the bacterial peptidoglycan products MDP and Tri-DAP by hPHT1. In doing so, the proposed studies may offer significant insight into the molecular mechanism of PHT1-mediated bacterial peptidoglycan-induced inflammation in cells, and into PHT1 being used as a potential drug target for reducing the immune response and, thereby, treating inflammatory diseases.
2. MATERIALS AND METHODS
2.1. Materials
d3-l-histidine was purchased from PUEN Scientific Instrument Corporation (Guangzhou, China). MDP and Tri-DAP were purchased from InvivoGen (San Diego, CA, USA). Primers used in the present study were synthesized by Sangon Biotech Corporation (Shanghai, China). Specific polyclonal antibody for GFP was obtained from Sangon (Shanghai, China). The monoclonal antibody for GAPDH, and secondary goat antirabbit, and goat antimouse antibodies were purchased from Multisciences Biotech Corporation (Hangzhou, China). Lipofectamine 3000 and hygromycin B were obtained from Invitrogen (Carlsbad, CA, USA). GlySar was purchased from Sigma-Aldrich (St. Louis, MO, USA) and all other chemicals were obtained from standard sources.
2.2. hPHT1-EGFP Plasmid Construction
The cDNA of wildtype hPHT1 was cloned by proofreading PCR using the reverse transcripts of human THP-1 cells total RNA. The primers were designed as follows: forward primer 5′-CGCAAGCTTCG-CATGGAGGGCTCTG-3′ (containing an artificially introduced Hind III site) and reverse primer 5′-AATGGATCCGGCCCT-CCTGCTGGTG-3′ (containing an artificially introduced BamH I site that removes stop codon). The PCR product was digested by restriction enzymes (ThermoFisher Scientific, Waltham, MA, USA), purified, and ligated into the Hind III/BamH I sites of pcDNA3.1-(+)-EGFP. The pcDNA3.1-(+)-EGFP plasmid was reconstructed by inserting the full-length EGFP cDNA into pcDNA3.1-(+) (Invitrogen, USA) at EcoR V and Xhol I sites. The sequence of the entire hPHT1 gene was confirmed by DNA sequencing core (Sangon, China).
2.3. Site-Directed Mutagenesis
Site-directed mutagenesis of the hPHT1-EGFP plasmid was constructed using PrimeStar HS DNA polymerase Kit (Takara, Japan) with the mutant primers listed in Table S1. The resulting hPHT1-(L14A, L15A)-EGFP and hPHT1 (L318A, V319A)-EGFP plasmid DNAs were isolated using the QIAprep Spin Miniprep Kit (Qiagen, Germany) and verified by sequence analysis. Once positive mutant colonies were obtained, plasmid mutant DNA was used for the second mutagenesis to construct hPHT1-(L14A, L15A, L318A, V319A)-EGFP plasmids, which were then confirmed by sequencing the mutants.
2.4. Cell Culture and Transfection
For stable transfection, MDCK cells (Type Culture Collection of the Chinese Academy Sciences, Shanghai, China) were cultured in DMEM medium (Invitrogen, USA) supplemented with 10% FBS (Sciencell, Carlsbad, CA, USA), 1% penicillin, streptomycin at 37 °C in a 5% CO2 incubator. Lipofectamine 3000 was used for delivery of insertion constructs pcDNA3.1-(+)-EGFP, pcDNA3.1-(+)-hPHT1WT-EGFP, and pcDNA3.1-(+)-hPHT1mut-EGFP into MDCK cells. After transfection for 48 h, selection medium containing 400 µg/mL hygromycin B was added and maintained for 7 days. Hygromycin B resistant colonies were identified by fluorescence for further analysis. For transient transfection, Hela cells were cultured 24 h prior to transfection in Nunc glass bottom dishes (ThermoFisher Scientific, USA) and hPHT1-EGFP, hPHT1-(L14A, L15A)-EGFP, hPHT1-(L318A, V319A)-EGFP, and hPHT1-(L14A, L15A, L318A, V319A)-EGFP plasmids were inserted with Lipofectamine 3000 when the density reached 2 × 106 cells/ml. After 24 h, the cells were analyzed under the confocal laser scanning fluorescence microscope (FV3000; Olympus, Japan) with an oil immersion lens (×60 magnification, NA 1.42).
2.5. mRNA Analysis
Total RNAs were extracted from MDCK-EGFP (mock) and MDCK-hPHT1mut-EGFP (hPHT1mut) cells (Axygen, Tewksbury, MA, USA) and the cDNAs generated with PrimeScript RT reagent kit (Takara, Japan). The cDNAs were then amplified using SYBR Premix Ex TaqII (Takara, Japan) and detected by qRT-PCR (Applied Biosystems StepOnePlus, USA). mRNA expression of target genes was normalized to the house-keeping gene Gapdh and described as 2−ΔCt, ΔCt = average Ct (target genes) – average Ct (Gapdh). Primers for qRT-PCR were listed in Table 1.
Table 1.
qRT-PCR Primers for Target Genes in hPHT1mut and Mock Cells
| gene | forward primer (5′-3′) | reverse primer (5′-3′) |
|---|---|---|
| Lat1 | CACCCATGCCTTCCCTTGT | GTCTCTGGAGAAGGCGTAAAGC |
| Ntt4 | CATCCCTCCCCACGTCAAC | TGACTCTGTACATGTCTGTGTAGTCCTT |
| Pat1 | TGATAGAAGCGGCCAATGG | GCGTCAGAATCACCGTCTCA |
| Pht1 | GCGTGGCTTTCCTGGTCTT | ATCGGGAGGCTTGGTGATAA |
| Snat1 | AAGGAAACAGGCTGCATGGT | TCCCTGTGGTGCCAAAGAC |
| Snat3 | CAGCTTGGCTACCTGGGTTACT | AGGAAGAACACCATGCAACTGA |
| hPHT1 | ACCACTCCTCACACGCTCCCTG | GGAGGATGGGAGCAGGCCAT |
| Gapdh | GCACCGTCAAGGCTGAGAAC | TGGTGAAGACGCCAGTGGA |
2.6. Immunoblotting of hPHT1-GFP
Protein samples were collected from stably transfected MDCK cells, lysed with Nonidet P40-lysis buffer (50 mM Tris-HCl, 0.4 M NaCl, 0.5% NP-40, 10% glycerol, 1 mM EDTA, pH 8.0), and then centrifuged at 13 000 g × 15 min at 4 °C. Denatured samples were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (Bio-Rad, Hercules, CA, USA) and transferred onto nitrocellulose membranes (Millipore Corporation, Billerica, MA, USA). The membrane was blocked at room temperature for 3 h with 5% nonfat milk in Tris-buffered saline with 0.1% Tween 20 (TBST), washed with TBST for 3 min, 3 times, and then incubated with the primary anti-GFP antibody (1:1000) or anti-GAPDH antibody (1:10000) overnight at 4 °C. The membranes were washed three times and incubated with a second antibody, goat antirabbit or goat antimouse IgG-horseradish peroxidase (1:5000) for 2 h at room temperature. After washing with TBST, three times, the bound antibody was incubated with ECL Western Blotting Substrate (GE Healthcare, Pittsburgh, PA, USA) and detected by the Alpha FluorChem E System (ProteinSimple, San Jose, CA, USA).
2.7. Uptake Studies
Uptake studies were performed with d3-l-histidine, GlySar, MDP, or Tri-DAP using a method described previously.5,27 After 24 h of culture, stably transfected cells were washed twice with MES buffer (8 g NaCl, 0.4 g KCl, 0.14 g CaCl2, 0.2 g MgSO4·7H2O, 0.06 g Na2HPO4·12H2O, 0.06 g KH2PO4, 0.35 g NaHCO3, 1 g glucose, 1.95 g MES in 1 L ddH2O, pH 6.5) at 37 °C. Uptake was initiated by incubating the cells with MES buffer containing specific concentrations of d3-l-histidine, GlySar, MDP, or Tri-DAP for designated periods of time. For time-dependent uptake, cells were incubated for 5, 10, 20, and 30 min. For pH-dependent studies, the uptake was evaluated over a pH range of 4.5 to 7.4. For concentration-dependent studies, d3-l-histidine and GlySar were determined over the concentration range of 1 to 1000 µM and 20 to 2000 µM, respectively, using 15 min incubations. Inhibition studies were conducted with 10 µg/mL MDP in the absence or presence of potential inhibitors (2 mM) or indicated concentrations of histidine or GlySar (0.01–50 mM) and incubated for 30 min. All uptake studies were terminated by removing the buffer and rapidly washing three times with ice-cold MES buffer. The cells were then treated with 0.1% SDS and the concentrations were determined by HPLC-MS/MS (Agilent 6460, Agilent, Santa Clara, CA, USA). Values were corrected for cell number variations by quantifying protein content using the Bicinchoninic acid assay kit (Beyotime, China).
2.8. HPLC-MS/MS Detection
In the uptake studies, all the compounds were detected by an Agilent 6460 triple quadrupole mass spectrometer with an ESI source in positive ion mode, equipped with an Agilent 1290 liquid chromatography system. The chromatographic separation was performed on an ZORBAX SB-AQ column (5 µm, 4.6 × 150 mm; Agilent) maintained at 35 °C with a mobile phase flow rate of 0.6 mL/min, where mobile phase A consisted of water with 20 mM ammonium and mobile phase B consisted of acetonitrile with 0.1% formic acid. Quantification was obtained using multiple reaction monitoring (MRM) mode, the m/z transitions of all the compounds were listed in Table S2. The mass spectrometer parameters were optimized as follows: gas temperature 325 °C, gas flow 5 L/min, nebulizer 45 psi, capillary voltage 3500 V, sheath gas temperature 350 °C, and sheath gas flow 11 L/min. Agilent MassHunter software (version B.04.01; Agilent) was used for data acquisition and analysis.
2.9. Data Analysis
Data are expressed as mean ± SE of three independent experiments with each experiment being carried out in triplicate. Concentration-dependent cellular uptake of d3-l-histidine and GlySar were best fitted to a Michaelis– Menten equation:
Where V represents the cellular uptake rate, Vmax the maximum uptake rate, Km the Michaelis constant, and C the substrate (d3-l-histidine or GlySar) concentration, after being corrected for uptake in the mock cells.
A comparison between two treatment groups was performed by an unpaired t test and among multiple treatment groups using one-way analysis of variance (ANOVA) followed by the Dunnett’s test (GraphPad Prism, v6.0; GraphPad Software, Inc. c., La Jolla, CA, USA). Values of p ≤ 0.05 were considered to be statistically significant.
3. RESULTS
3.1. Mutation of Two Dileucine Motifs Localize hPHT1 to Plasma Membrane
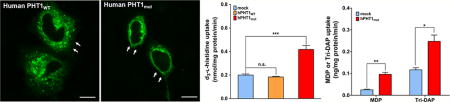
To elucidate the characteristics of wildtype PHT1 is difficult because PHT1 is localized in the membranes of endosomes and lysosomes, and model substrates are required to cross the extracellular membranes first. To overcome this technical challenge, three novel hPHT1 mutants were constructed and evaluated whether they were localized in the plasma membrane by immunofluorescence microscopy. As shown in Figure 1, human, mouse, and rat PHT1 had two dileucine motifs (EXXXLL/DXXXLV) in their protein sequences. In human, one dileucine motif was presented in the N-terminal at amino acids 14 and 15 and the other in T7 at amino acids 318 and 319 (Figure 1A and B). When the first of two dileucine motifs was substituted by alanine, hPHT1 was still localized in the membrane of lysosomes. Likewise, when the second of two dileucine motifs was replaced by alanine, no change was observed in the subcellular location of PHT1. However, when both dileucine motifs were substituted by alanine, hPHT1 was localized to the plasma membrane (Figure 1C). To compare the transport activity of wildtype and mutant hPHT1, the uptake of 10 µM histidine was evaluated in MDCK cells stably transfected with hPHT1WT and hPHT1mut. As shown in Figure 1D, the uptake of histidine in hPHT1mut cells was 2-fold greater than that of mock cells, whereas no significant difference was observed in hPHT1WT as compared to mock cells.
Figure 1.
Mutation of two dileucine motifs localize hPHT1 to plasma membrane. (A) The signal pathway of hPHT1 expression. Wildtype hPHT1 protein was targeted to express in the membrane of endosomes and lysosomes. However, mutation of two dileucine-based motifs resulted in hPHT1 localizing to plasma membranes. The hPHT1 putative protein was predicted to contain 577 amino acids and 12 transmembrane domains (T1-T12) with the N- and C-termini in cytosol. (B) Dileucine motifs in mammalian PHT1. Human, mouse, and rat PHT1 have two dileucine motifs ([E/D]-xxxLL/LV). (C) Fluorescence microscopy of the dileucine mutants of hPHT1-EGFP in Hela cells. Either of the two dileucine motifs substituted by alanine was insufficient to localize the protein to plasma membranes. Cell membranes are marked by arrows. Bars, 10 µm. (D) MDCK cells stably transfected with EGFP (mock), hPHT1WT, and hPHT1mut plasmids were incubated with 10 µM d3-l-histidine for 15 min. Data are expressed as mean ± SE(n = 3); n.s., not significant; ***p < 0.001, as compared to mock cells.
3.2. Expression and Functional Characterization of hPHT1mut
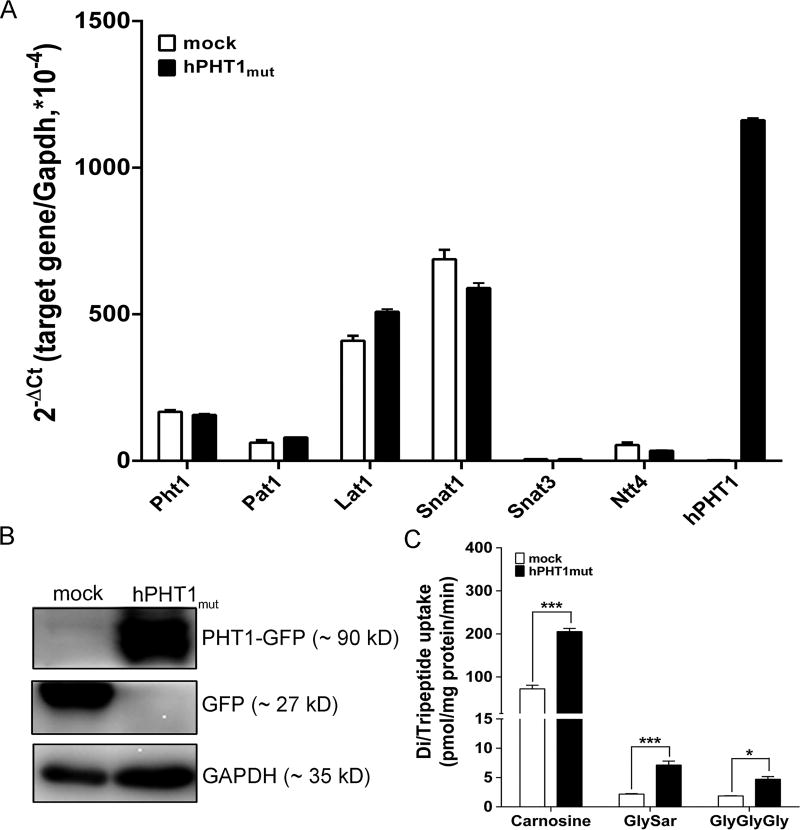
The mRNA expression of endogenous canine Pht1, heterologous hPHT1 and other amino acid transporters which probably transport histidine were determined in hPHT1mut and mock cells. The results showed that endogenous Pht1 was very close in both cell systems at very low levels, and heterologous hPHT1 mRNA expression was substantially higher in hPHT1mut than mock cells (Figure 2A). Since no suitable hPHT1 antibody was available, an antibody was raised against GFP to determine the protein expression of hPHT1. Only one band was found for GFP in mock cells (~27 kDa), whereas one band was found for PHT1-GFP in hPHT1mut cells (~90 kDa), indicating that hPHT1 was about 63 kDa (Figure 2B). As PHT1 mediates the transport of di/tripeptides and histidine, the cellular uptake of typical substrates was measured in hPHT1mut and mock cells. As observed in Figure 2C, the uptakes of carnosine, GlySar and GlyGlyGly were 2- to 4-fold greater in hPHT1mut cells.
Figure 2.
Expression and functional characterization of hPHT1mut. (A) mRNA expression of endogenous canine Pht1, heterologous hPHT1, and amino acid transporters in hPHT1mut and mock cells. (B) Protein expression of GFP in hPHT1mut and mock cells. (C) Uptake of l-carnosine, GlySar, and GlyGlyGly in hPHT1mut and mock cells. Uptake studies were performed over 15 min with 100 µM l-carnosine, 100 µM GlySar and 100 µM GlyGlyGly in MES buffer (pH 6.5). Data are expressed as mean ± SE (n = 3). Statistical analyses were performed using an unpaired t test, where *p < 0.05, **p < 0.01, and ***p < 0.001, as compared to mock cells.
3.3. Uptake Kinetics of d3-l-Histidine in hPHT1mut Cells
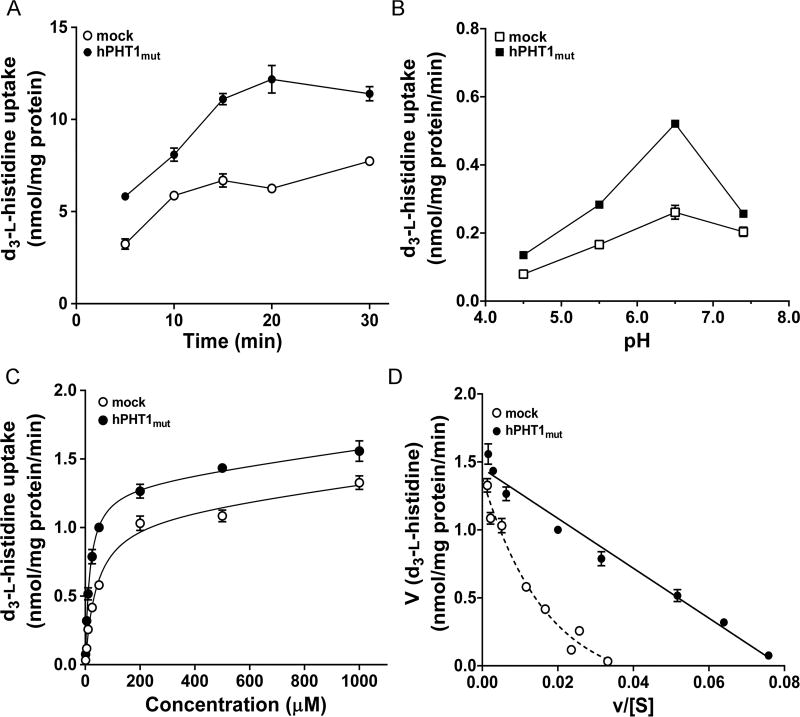
Our results revealed that the uptake of d3-l-histidine in hPHT1mut cells was time and pH-dependent (Figure 3A and B). Since uptake was linear over 15 min and optimized at a buffer pH of 6.5, these values were selected for the subsequent concentration-dependent study. As shown in Figure 3C, hPHT1mut and mock cells displayed a saturable uptake of d3-l-histidine, which were fit to a Michaelis–Menten term. The Km value of hPHT1mut for d3-l-histidine was 16.3 ± 1.9 µM and the Vmax was 1317 ± 51 pmol/mg protein/min (Figure 3C and Table 2). The transport rates mediated by hPHT1mut or endogenous transporters were transformed according to Eadie-Hofstee, and the resultant plot for hPHT1mut shown to be linear (r2 = 0.9661) (Figure 3D), while for the latter was nonlinear.
Figure 3.
Uptake kinetics of d3-l-histidine in hPHT1mut cells. (A) Time-dependent uptake of 10 µM d3-l-histidine in hPHT1mut and mock cells at pH 6.5. (B) pH-dependent uptake of 10 µM d3-l-histidine in hPHT1mut and mock cells after 15 min incubations. (C) Concentration-dependent uptake of d3-l-histidine (1–1000 µM) in hPHT1mut and mock cells after 15 min incubations in MES buffer (pH 6.5). Uptakes of d3-l-histidine in mock cells were subtracted from the corresponding uptakes in MDCK-hPHT1mut cells. (D) The transport rates were transformed according to Eadie–Hofstee. Data are expressed as mean ± SE (n = 3).
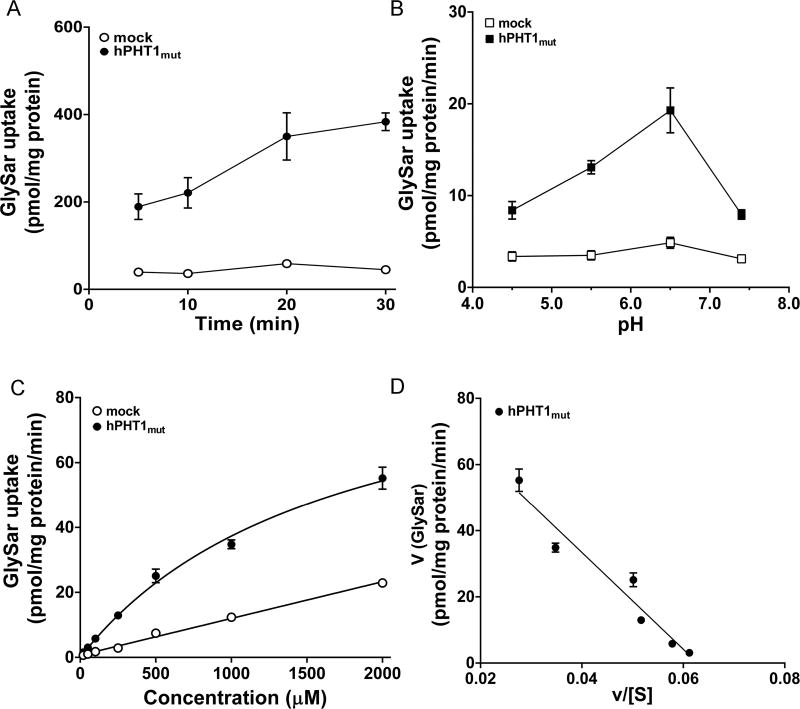
Table 2.
Transport Kinetics of d3-l-histidine and GlySar Uptake in MDCK Cells Expressing hPHT1muta
| parameters (units) | d3-l-histidine | GlySar |
|---|---|---|
| Vmax (pmol/mg protein/min) | 1317 ± 51 | 100 ± 10 |
| Km (µM) | 16.3 ± 1.9 | 1690 ± 300 |
| Vmax/Km (µL/mg protein/min) | 80.8 | 0.0592 |
| r2 | 0.985 | 0.979 |
Data are expressed as mean ± SE (n = 3).
3.4. Concentration-Dependent Uptake of GlySar
GlySar uptake was substantially greater in hPHT1mut cells than mock cells (Figure 2C). However, the affinity of hPHT1 for GlySar is unknown. Therefore, the concentration-dependent uptake of GlySar was evaluated in hPHT1mut cells. As before, time and pH-dependent studies were performed to determine the optimal incubation conditions for GlySar transport. As shown in Figure 4A and B, an incubation time of 15 min and buffer pH of 6.5 were considered optimal for uptake. The data revealed that hPHT1mut demonstrated a saturable transport profile of GlySar, which was fit to a single Michaelis–Menten term and then transformed to a Eadie–Hofstee plot (Figure 4C and D). The estimated Km and Vmax values were 1.69 ± 0.30 mM, and 100 ± 10 pmol/mg protein/min, respectively (Table 2).
Figure 4.
Concentration-dependent uptake of GlySar in hPHT1mut cells. (A) Time-dependent uptake of 100 µM GlySar in hPHT1mut and mock cells at pH 6.5. (B) pH-dependent uptake of 100 µM GlySar in hPHT1mut and mock cells after 15 min incubations. (C) Concentration-dependent uptake of GlySar (20–2000 µM) in hPHT1mut and mock cells after 15 min incubations in MES buffer (pH 6.5). Uptakes of GlySar in mock cells were subtracted from the corresponding uptakes in hPHT1mut cells. (D) Eadie–Hofstee plot of GlySar uptake. Data are expressed as mean ± SE (n = 3).
3.5. MDP and Tri-DAP Are Substrates of hPHT1mut
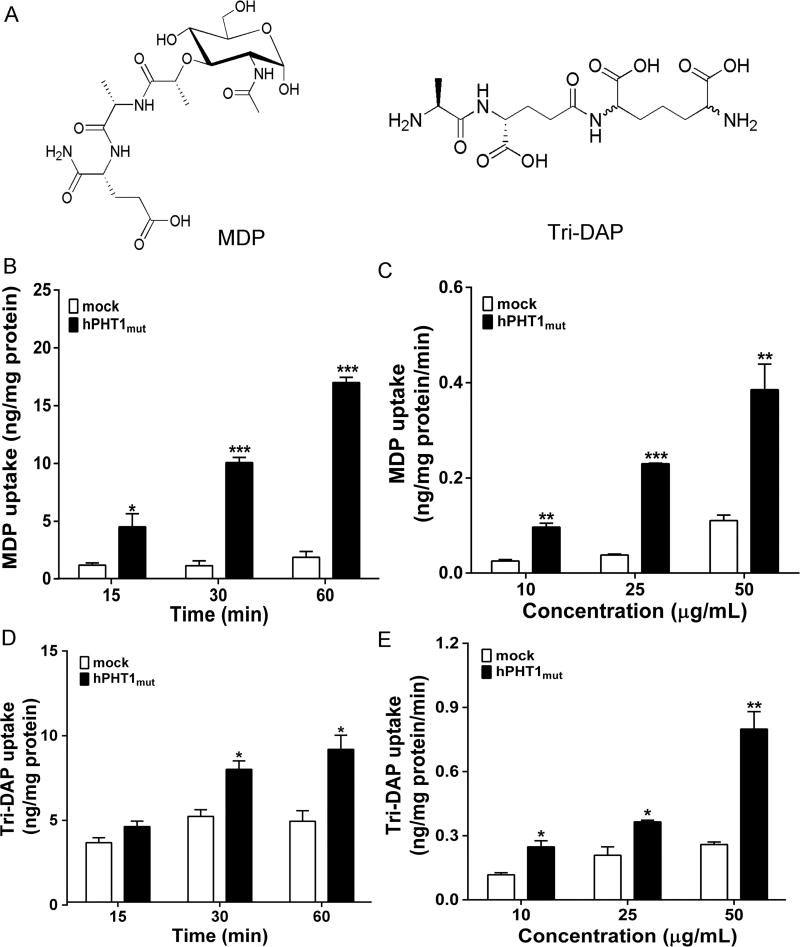
PHT1 was postulated as being responsible for the peptidoglycan-induced immune responses since di/tripeptide peptidoglycans (e.g., MDP as NOD2 ligand and Tri-DAP as NOD1 ligand) might be substrates of hPHT1. To further test this postulation, hPHT1mut and mock cells were incubated with 10 µg/mL MDP or Tri-DAP over a 60 min period. As shown in Figure 5B and D, the uptakes of MDP and Tri-DAP were 8-fold and 2-fold greater, respectively, in hPHT1mut cells than that observed in mock cells at 60 min. Figure 5C and E demonstrated that the 30 min cellular uptakes of MDP and Tri-DAP increased with increasing substrate concentrations (i.e., at 10, 25, and 50 µg/mL or about 20–100 µM) and that the uptakes in hPHT1mut cells were significantly higher at each concentration, as compared to mock cells.
Figure 5.
MDP and Tri-DAP are substrates of hPHT1mut. (A) Chemical structures of MDP and Tri-DAP. (B and D) Time-dependent uptake of 10 µg/mL MDP (B) and Tri-DAP (D) in hPHT1mut and mock cells at buffer pH 6.5. (C and E) Concentration-dependent uptake of MDP (C) and Tri-DAP (E) in hPHT1mut and mock cells, as performed over 30 min incubations in MES buffer (pH 6.5). Data are expressed as mean ± SE (n = 3). Statistical analyses were performed using an unpaired t test, where *p < 0.05, **p < 0.01, and ***p < 0.001, as compared to mock values.
3.6. Inhibition of MDP Uptake in hPHT1mut Cells
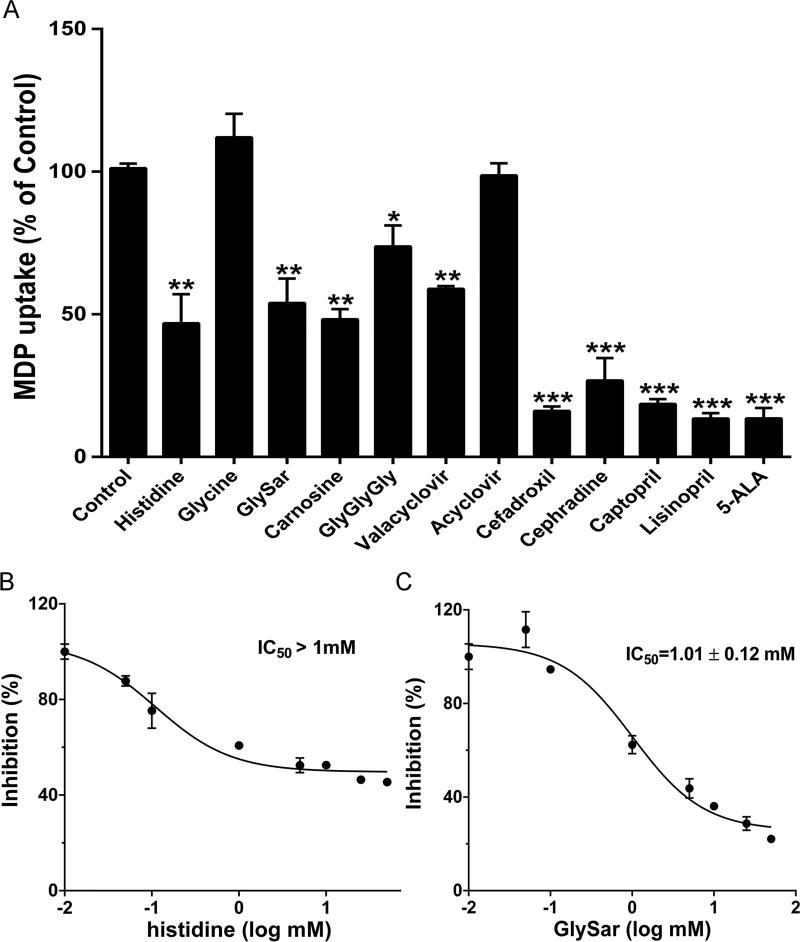
The specificity of MDP uptake was evaluated in hPHT1mut cells in the absence and presence of 2 mM potential inhibitors, such as amino acids, di/tripeptides, antiviral drugs (valacyclovir and acyclovir), cephalosporins with an α-amino group, ACE inhibitors, and 5-ALA (Figure 6A). The amino acid glycine had no effect on the hPHT1-mediated uptake of MDP, whereas histidine reduced MDP uptake to less than 50% of control. As expected, the uptake of MDP was inhibited by 50% in the presence of dipeptides (GlySar or carnosine) and by 25% in the presence of a tripeptide GlyGlyGly. With respect to peptidomimetics, the antiviral prodrug valacyclovir reduced MDP uptake by 45%, whereas its active moiety acyclovir was without effect. Finally, the uptake of MDP was suppressed by 70–85% in the presence of aminocephalosporins (cefadroxil or cephradine), ACE inhibitors (captopril or lisinopril), and 5-ALA. Since the affinity of histidine was much higher than GlySar, but they had similar inhibitory effects, the dose–response studies were performed. As shown in Figure 6B and C, the IC50 value is higher than 1.0 mM for histidine (i.e., much different than Km value) and 1.0 mM for GlySar (i.e., similar to the Km value).
Figure 6.
Inhibition of MDP uptake in hPHT1mut cells. (A) Effect of test compounds on the inhibition of MDP uptake in hPHT1mut cells. (B and C) Uptake of MDP into hPHT1mut cells as a function of increasing concentration of histidine (B) and GlySar (C). Uptake studies were performed with 10 µg/mL MDP in hPHT1mut and mock cells in the absence or presence of 2 mM potential inhibitors for (A) or 0.01 to 50 mM of histidine or GlySar for (B) and (C) within 30 min incubations (buffer pH 6.5). The uptakes in mock cells were subtracted from the corresponding uptakes in hPHT1mut cells. Data are expressed as mean ± SE (n = 3). Statistical analyses were performed using oneway ANOVA followed by Dunnett’s test, where *p < 0.05, **p < 0.01, and ***p < 0.001, as compared to control values.
4. DISCUSSION
There is a paucity of data on the functional activity of hPHT1 substrates, such as histidine, di/tripeptides, and peptide-like drugs. Previous uptake experiments in Xenopus laevis oocytes have demonstrated that rat PHT1 was a high affinity transporter for histidine and the transport process was protondependent.8 Moreover, in vitro brain slice studies showed that histidine reduced the uptake of GlySar by 81% in adult rat, indicating PHT1 plays a dominant role in GlySar brain uptake.28 However, these studies did not evaluate the substrate spectrum of hPHT1 or the uptake kinetics of hPHT1 for histidine and other di/tripeptides.
In the present study, we reported several new findings regarding the functional activity of hPHT1 in stably transfected MDCK cells. We found that (1) mutation of two dileucine-based motifs was essential for hPHT1 to preferentially localize to plasma membranes, and the plasma transporter was responsible for the intracellular accumulation of histidine, carnosine, GlySar and GlyGlyGly; (2) hPHT1-mediated transport of histidine and GlySar was saturable, and the affinity of hPHT1 for histidine (Km = 16.3 µM) was much higher than that of GlySar (Km = 1.69 mM); and (3) MDP and Tri-DAP were substrates of hPHT1 and the uptake of MDP was inhibited by histidine, di/tripeptides, and peptide-like drugs.
Histidine is one of the proteinogenic amino acids and possesses many crucial biological activities, such as detoxification of heavy metals and the manufacturing of white and red blood cells.29 Histidine is also a precursor of histamine, carnosine, ergothioneine and vitamin C.30 The availability of histidine is positively correlated with its production in brain.31,32 Moreover, in several neurological diseases, such as multiple sclerosis, Alzheimer’s disease, Down’s syndrome, and Wernicke’s encephalopathy, the histamine levels in brain were significantly changed.33 Several transporters were responsible for transport of histidine in the central nervous system, including the SLC38 (SNATs, Na+-dependent), SLC6 (e.g., NTT4, sodium and chloride-dependent), SLC7 (e.g., LATs, Na+-independent) families, and peptide/histidine transporters (e.g., PHT1 and PHT2).28,34 However, little information is known about the relative importance of amino acid transporters and PHT1 in this process. A recent study showed that the uptake of histidine was reduced in brain slices by 50% during PHT1 ablation and the amino acid transporters accounted for 30% of the uptake.35 In our concentration-dependent studies, we found that hPHT1 was a high affinity transporter for histidine (Km = 16.3 ± 1.9 µM), which was consistent with the result (Km = 17 µM) in rPHT1-transfected Xenopus laevis oocytes.8
In the uptake study, we found no difference in the uptake of histidine in mock and hPHT1WT cells. However, other authors showed that the uptake of histidine was different when expressing wildtype PHT1 in Xenopus laevis oocytes or COS-7 cells.8,22,36 Although speculative, this “apparent” inconsistency might be due to the different expression system being studied (i.e., amphibian vs mammalian) as well as the different method of transfection (i.e., transient vs stable). The latter point may explain why the same group found no uptake of GlySar in COS-7 cells transiently transfected with hPHT1,22 whereas GlySar uptake was observed in COS-7 cells stably transfected with hPHT1.36 Additionally, our results showed that mRNA expression of Lat1 and Snat1 were much higher than endogenous Pht1, which could explain the transporters-mediated uptake of histidine in mock cells (Figure 2A, Figure 3C and D). Furthermore, Sasawatari et al. found that histidine could suppress the production of TLR9-dependent proinflammatory cytokines in dendritric cells.13 Similarly, Andou et al. reported that histidine could ameliorate murine colitis by inhibition of NF-κB activation and down-regulation of proinflammatory cytokine production.37 These findings suggest that PHT1 may play an important role in histidine transport and inflammatory responses, as well as in endogenous histidine/histamine homeostasis and immune responses. Further studies are needed to elucidate the molecular mechanism of PHT1-mediated transport of lysosomal histidine to control immune responses.
GlySar is a synthetic dipeptide typically used as a model substrate for PEPT1 and PEPT2. However, PEPT1 and PEPT2 show opposite affinities for GlySar, with mM values of Km for PEPT1 and µM values of Km for PEPT2.38,39 Our kinetic analysis of GlySar uptake indicated hPHT1 was a low affinity transporter (Km = 1.69 ± 0.30 mM), whose transport activity was similar to PEPT1. However, Bhardwaj et al. reported that the uptake of GlySar was not different in COS-7 transiently transfected with wildtype hPHT1 or vector.22 Their inhibition study showed that the uptake of GlySar in retinal epithelial cell lines was also not affected by histidine,22 which can be attribute to the low passive diffusion of GlySar and the subcellular location of PHT1 in mammalian cells.
Studies on the functional relevance of PHT1 in TLR- and NOD-induced inflammatory responses suggest that PHT1 might mediate the transport of MDP, Tri-DAP or γ-iE-DAP. A recent study showed that the PHT1-mediated efflux of MDP from endosomes was responsible for the activation of NOD2-dependent signals, as obtained using NF-κB reporter assays.21 However, there is no direct evidence to clarify whether MDP or Tri-DAP are substrates of PHT1. Sasawatari et al. proposed that PHT1 was not a MDP transporter based on the finding that no difference was observed in the expression of proinflammatory cytokines in MDP-treated SLC15A4 wildtype and deficient mice.13 These contrary results indicated that a robust experimental model was needed to demonstrate whether MDP and Tri-DAP were substrates of PHT1. In the present study, we proved MDP and Tri-DAP were substrates of PHT1 using the MDCK cell stably transfected system hPHT1mut. Still, it was not easy to estimate the affinity of hPHT1 for both substrates (i.e., Km) because of the low solubility of MDP and Tri-DAP.
Since MDP transport is related to immune responses, inhibiting the PHT1-mediated uptake of MDP may be an effective strategy to control immune responses. Thus, we evaluated several potential inhibitors of MDP uptake and found that peptide-like drugs had a more potent inhibitory effect than histidine and di/tripeptides (Figure 6A). The inhibition of MDP uptake by di/tripeptides (25–50%) was in accordance with the results observed previously in histidine inhibition studies.8,22 However, the IC50 value of histidine was much different with its Km value, but GlySar was nearly the same (Figure 6B and C). Because histidine could be transported by several transporters, we supposed initially the discrepancy might be attributed to the low concentration of histidine in the extracellular medium. With this in mind, we determined the concentration of histidine in cell medium after the uptake study was terminated (i.e., at both 1.0 and 5.0 mM), but the extracellular concentrations of histidine were close to the indicated concentrations (data not shown). The discrepancy between Km and IC50 values for histidine was difficult to explain, but we would further investigate this issue in subsequent studies.
Several studies have suggested that PHT1 mutations are associated with the susceptibility of systemic lupus erythematosus,9,10,12 while Baccala et al. proposed that PHT1 could be a candidate target for human lupus therapy.40 The physiological, pharmacological, and pathological relevance of PHT1 still needs to be fully characterized. Our novel approach in targeting and expressing hPHT1 in plasma membranes might be a useful tool for describing substrate structural properties and drug design for hPHT1 in the future.
Taken together, our study characterized a novel hPHT1 mutant model that expressed hPHT1 in plasma membranes by disruption of two dileucine motifs. hPHT1 showed high affinity for histidine but low affinity for GlySar, and the uptake of both substrates was pH dependent. Bacterial peptidoglycan components, such as MDP and Tri-DAP, were substrates of hPHT1 and their uptake was inhibited by di/tripeptides and peptide-mimetics. It appears that hPHT1 may be a potential target for reducing immune responses and drug treatment of inflammatory diseases. The stably transfected hPHT1-MDCK cell model could be a useful tool to study potential substrates or inhibitors of hPHT1, which would provide a valuable bridge in better understanding the role of PHT1 in physiological and pathological states.
Supplementary Material
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant Nos. 81573492), the Zhejiang Provincial Science and Technology Foundation of China (2015C33162), and the National Institutes of Health (Grant R01 GM115481 to DES).
ABBREVIATIONS USED
- ACE
angiotensin converting enzyme
- 5-ALA
5-aminolevulinic acid
- GlyGlyGly
glycylglycyl-glycine
- GlySar
glycylsarcosine
- hPEPT1
human PEPT1 transporter
- hPEPT2
human PEPT2 transporter
- hPHT1
human PHT1 transporter
- hPHT2
human PHT2 transporter
- HPLC-MS/MS
high-performance liquid chromatography–mass spectrometer
- iE-DAP
γ-d-glutamyl-meso-diaminopimelic
- MDP
muramyl peptide
- MTri-DAP
N-acetylmuramyl-l-Ala-d-Glu-mesoDAP
- NF-κB
nuclear factor-kappa B
- NOD1
nucleotide-binding oligomerization domain 1
- NOD2
nucleotide-binding oligomerization domain 2
- PCR
polymerase chain reaction
- RIPK2
serine/threonine-protein kinase 2
- SLC15A
solute carrier family 15 A
- TLRs
toll-like receptors
- Tri-DAP
l-Ala-γ-d-Glu-meso-DAP
Footnotes
ASSOCIATED CONTENT
- The m/z transitions of all the detected compounds, primers for site-directed mutagenesis (PDF)
The authors declare no competing financial interest.
References
- 1.Fei YJ, Ganapathy V, Leibach FH. Molecular and structural features of the proton-coupled oligopeptide transporter superfamily. Prog. Nucleic Acid Res Mol Biol. 1997;58:239–261. doi: 10.1016/s0079-6603(08)60038-0. [DOI] [PubMed] [Google Scholar]
- 2.Smith DE, Clemencon B, Hediger MA. Proton-coupled oligopeptide transporter family SLC15: Physiological, pharmacological and pathological implications. Mol. Aspects Med. 2013;34(2–3):323–336. doi: 10.1016/j.mam.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Daniel H, Kottra G. The proton oligopeptide cotransporter family SLC15 in physiology and pharmacology. Pfluegers Arch. 2004;447(5):610–618. doi: 10.1007/s00424-003-1101-4. [DOI] [PubMed] [Google Scholar]
- 4.Steiner HY, Naider F, Becker JM. The PTR family: a new group of peptide transporters. Mol. Microbiol. 1995;16(5):825–834. doi: 10.1111/j.1365-2958.1995.tb02310.x. [DOI] [PubMed] [Google Scholar]
- 5.Herrera-Ruiz D, Faria TN, Bhardwaj RK, Timoszyk J, Gudmundsson OS, Moench P, Wall DA, Smith RL, Knipp GT. A novel hPepT1 stably transfected cell line: establishing a correlation between expression and function. Mol. Pharmaceutics. 2004;1(2):136–144. doi: 10.1021/mp034011l. [DOI] [PubMed] [Google Scholar]
- 6.Rubio-Aliaga I, Daniel H. Peptide transporters and their roles in physiological processes and drug disposition. Xenobiotica. 2008;38(7–8):1022–1042. doi: 10.1080/00498250701875254. [DOI] [PubMed] [Google Scholar]
- 7.Sakata K, Yamashita T, Maeda M, Moriyama Y, Shimada S, Tohyama M. Cloning of a lymphatic peptide/histidine transporter. Biochem. J. 2001;356:53–60. doi: 10.1042/0264-6021:3560053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamashita T, Shimada S, Guo W, Sato K, Kohmura E, Hayakawa T, Takagi T, Tohyama M. Cloning and functional expression of a brain peptide/histidine transporter. J. Biol. Chem. 1997;272(15):10205–10211. doi: 10.1074/jbc.272.15.10205. [DOI] [PubMed] [Google Scholar]
- 9.He CF, Liu YS, Cheng YL, Gao JP, Pan TM, Han JW, Quan C, Sun LD, Zheng HF, Zuo XB, Xu SX, Sheng YJ, Yao S, Hu WL, Li Y, Yu ZY, Yin XY, Zhang XJ, Cui Y, Yang S. TNIP1, SLC15A4, ETS1, RasGRP3 and IKZF1 are associated with clinical features of systemic lupus erythematosus in a Chinese Han population. Lupus. 2010;19(10):1181–1186. doi: 10.1177/0961203310367918. [DOI] [PubMed] [Google Scholar]
- 10.Lee J, Tattoli I, Wojtal KA, Vavricka SR, Philpott DJ, Girardin SE. pH-dependent Internalization of Muramyl Peptides from Early Endosomes Enables Nod1 and Nod2 Signaling. J. Biol. Chem. 2009;284(35):23818–23829. doi: 10.1074/jbc.M109.033670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takeuchi F, Ochiai Y, Serizawa M, Yanai K, Kuzuya N, Kajio H, Honjo S, Takeda N, Kaburagi Y, Yasuda K, Shirasawa S, Sasazuki T, Kato N. Search for type 2 diabetes susceptibility genes on chromosomes 1q, 3q and 12q. J. Hum Genet. 2008;53(4):314–324. doi: 10.1007/s10038-008-0254-6. [DOI] [PubMed] [Google Scholar]
- 12.Zuo XB, Sheng YJ, Hu SJ, Gao JP, Li Y, Tang HY, Tang XF, Cheng H, Yin XY, Wen LL, Sun LD, Yang S, Cui Y, Zhang XJ. Variants in TNFSF4, TNFAIP3, TNIP1 BLK, SLC15A4 and UBE2L3 interact to confer risk of systemic lupus erythematosus in Chinese population. Rheumatol. Int. 2014;34(4):459–464. doi: 10.1007/s00296-013-2864-3. [DOI] [PubMed] [Google Scholar]
- 13.Sasawatari S, Okamura T, Kasumi E, Tanaka-Furuyama K, Yanobu-Takanashi R, Shirasawa S, Kato N, Toyama-Sorimachi N. The Solute Carrier Family 15A4 Regulates TLR9 and NOD1 Functions in the Innate Immune System and Promotes Colitis in Mice. Gastroenterology. 2011;140(5):1513–1525. doi: 10.1053/j.gastro.2011.01.041. [DOI] [PubMed] [Google Scholar]
- 14.Caruso R, Warner N, Inohara N, Nunez G. NOD1 and NOD2: Signaling, Host Defense, and Inflammatory Disease. Immunity. 2014;41(6):898–908. doi: 10.1016/j.immuni.2014.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Girardin SE, Boneca IG, Viala J, Chamaillard M, Labigne A, Thomas G, Philpott DJ, Sansonetti PJ. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J. Biol. Chem. 2003;278(11):8869–8872. doi: 10.1074/jbc.C200651200. [DOI] [PubMed] [Google Scholar]
- 16.Inohara N, Ogura Y, Fontalba A, Gutierrez O, Pons F, Crespo J, Fukase K, Inamura S, Kusumoto S, Hashimoto M, Foster SJ, Moran AP, Fernandez-Luna JL, Nunez G. Host recognition of bacterial muramyl dipeptide mediated through NOD2. J. Biol. Chem. 2003;278(8):5509–5512. doi: 10.1074/jbc.C200673200. [DOI] [PubMed] [Google Scholar]
- 17.Magalhaes JG, Philpott DJ, Nahori MA, Jehanno M, Fritz J, Le Bourhis L, Viala J, Hugot JP, Giovannini M, Bertin J, Lepoivre M, Mengin-Lecreulx D, Sansonetti PJ, Girardin SE. Murine Nod1 but not its human orthologue mediates innate immune detection of tracheal cytotoxin. EMBO Rep. 2005;6(12):1201–1207. doi: 10.1038/sj.embor.7400552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Opitz B, Puschel A, Schmeck B, Hocke AC, Rosseau S, Hammerschmidt S, Schumann RR, Suttorp N, Hippenstiel S. Nucleotide-binding oligomerization domain proteins are innate immune receptors for internalized Streptococcus pneumoniae. J. Biol. Chem. 2004;279(35):36426–36432. doi: 10.1074/jbc.M403861200. [DOI] [PubMed] [Google Scholar]
- 19.Charriere GM, Ip WKE, Dejardin S, Boyer L, Sokolovska A, Cappillino MP, Cherayil BJ, Podolsky DK, Kobayashi KS, Silverman N, Lacy-Hulbert A, Stuart LM. Identification of Drosophila Yin and PEPT2 as Evolutionarily Conserved Phagosome-associated Muramyl Dipeptide Transporters. J. Biol. Chem. 2010;285(26):20147–20154. doi: 10.1074/jbc.M110.115584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dalmasso G, Nguyen HTT, Charrier-Hisamuddin L, Yan Y, Laroui H, Demoulin B, Sitaraman SV, Merlin D. PepT1 mediates transport of the proinflammatory bacterial tripeptide L-Alagamma-D-Glu-meso-DAP in intestinal epithelial cells. Am. J Physiol-Gastr. L. 2010;299(3):G687–G696. doi: 10.1152/ajpgi.00527.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakamura N, Lill JR, Phung Q, Jiang ZS, Bakalarski C, de Maziere A, Klumperman J, Schlatter M, Delamarre L, Mellman I. Endosomes are specialized platforms for bacterial sensing and NOD2 signalling. Nature. 2014;509(7499):240–244. doi: 10.1038/nature13133. [DOI] [PubMed] [Google Scholar]
- 22.Bhardwaj RK, Herrera-Ruiz D, Eltoukhy N, Saad M, Knipp GT. The functional evaluation of human peptide/histidine transporter 1 (hPHT1) in transiently transfected COS-7 cells. Eur. J Pharm. Sci. 2006;27(5):533–542. doi: 10.1016/j.ejps.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 23.Bonifacino JS, Traub LM. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev Biochem. 2003;72:395–447. doi: 10.1146/annurev.biochem.72.121801.161800. [DOI] [PubMed] [Google Scholar]
- 24.Heilker R, Spiess M, Crottet P. Recognition of sorting signals by clathrin adaptors. BioEssays. 1999;21(7):558–567. doi: 10.1002/(SICI)1521-1878(199907)21:7<558::AID-BIES4>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 25.Botka CW, Wittig TW, Graul RC, Nielsen CU, Higaki K, Amidon GL, Sadee W. Human Proton/Oligopeptide transporter (POT) genes: Identification of putative human genes using bioinformatics. AAPS PharmSci. 2000;2(2):76–97. doi: 10.1208/ps020216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herrera-Ruiz D, Knipp GT. Current perspectives on established and putative mammalian oligopeptide transporters. J. Pharm. Sci. 2003;92(4):691–714. doi: 10.1002/jps.10303. [DOI] [PubMed] [Google Scholar]
- 27.Sun D, Wang Y, Tan F, Fang D, Hu Y, Smith DE, Jiang H. Functional and molecular expression of the proton-coupled oligopeptide transporters in spleen and macrophages from mouse and human. Mol. Pharmaceutics. 2013;10(4):1409–1416. doi: 10.1021/mp300700p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu YJ, Xie YH, Keep RF, Smith DE. Divergent developmental expression and function of the proton-coupled oligopeptide transporters PepT2 and PhT1 in regional brain slices of mouse and rat. J. Neurochem. 2014;129(6):955–965. doi: 10.1111/jnc.12687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kopple JD, Swendseid ME. Evidence That Histidine Is an Essential Amino-Acid in Normal and Chronically Uremic Man. J. Clin. Invest. 1975;55(5):881–891. doi: 10.1172/JCI108016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stifel FB, Herman RH. Histidine Metabolism. Am. J Clin. Nutr. 1971;24(2):207–217. doi: 10.1093/ajcn/24.2.207. [DOI] [PubMed] [Google Scholar]
- 31.Vaziri P, Dang K, Anderson GH. Evidence for histamine involvement in the effect of histidine loads on food and water intake in rats. J. Nutr. 1997;127(8):1519–1526. doi: 10.1093/jn/127.8.1519. [DOI] [PubMed] [Google Scholar]
- 32.Yoshimatsu H, Chiba S, Tajima D, Akehi Y, Sakata T. Histidine suppresses food intake through its conversion into neuronal histamine. Exp. Biol. Med. 2002;227(1):63–68. doi: 10.1177/153537020222700111. [DOI] [PubMed] [Google Scholar]
- 33.Haas HL, Sergeeva OA, Selbach O. Histamine in the nervous system. Physiol. Rev. 2008;88(3):1183–1241. doi: 10.1152/physrev.00043.2007. [DOI] [PubMed] [Google Scholar]
- 34.Schioth HB, Roshanbin S, Hagglund MGA, Fredriksson R. Evolutionary origin of amino acid transporter families SLC32, SLC36 and SLC38 and physiological, pathological and therapeutic aspects. Mol. Aspects Med. 2013;34(2–3):571–585. doi: 10.1016/j.mam.2012.07.012. [DOI] [PubMed] [Google Scholar]
- 35.Wang XX, Hu YJ, Keep RF, Toyama-Sorimachi N, Smith DE. A novel role for PHT1 in the disposition of L-histidine in brain: In vitro slice and in vivo pharmacokinetic studies in wildtype and Pht1 null mice. Biochem. Pharmacol. 2017;124:94–102. doi: 10.1016/j.bcp.2016.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lindley DJ, Carl SM, Mowery SA, Knipp GT. The Evaluation of Peptide/Histidine Transporter 1 (Pht1) Function: Uptake Kinetics Utilizing a Cos-7 Stably Transfected Cell Line. Rev. Mex Cienc. Farm. 2011;42(4):57–65. [PMC free article] [PubMed] [Google Scholar]
- 37.Andou A, Hisamatsu T, Okamoto S, Chinen H, Kamada N, Kobayashi T, Hashimoto M, Okutsu T, Shimbo K, Takeda T, Matsumoto H, Sato A, Ohtsu H, Suzuki M, Hibi T. Dietary Histidine Ameliorates Murine Colitis by Inhibition of Proinflammatory Cytokine Production From Macrophages. Gastroenterology. 2009;136(2):564–574. doi: 10.1053/j.gastro.2008.09.062. [DOI] [PubMed] [Google Scholar]
- 38.Hu YJ, Chen XM, Smith DE. Species-Dependent Uptake of Glycylsarcosine but Not Oseltamivir in Pichia pastoris Expressing the Rat, Mouse, and Human Intestinal Peptide Transporter PEPT1. Drug Metab. Dispos. 2012;40(7):1328–1335. doi: 10.1124/dmd.111.044263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Song FF, Hu YJ, Jiang HD, Smith DE. Species Differences in Human and Rodent PEPT2-Mediated Transport of Glycylsarcosine and Cefadroxil in Pichia Pastoris Transformants. Drug Metab. Dispos. 2017;45(2):130–136. doi: 10.1124/dmd.116.073320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baccala R, Gonzalez-Quintial R, Blasius AL, Rimann I, Ozato K, Kono DH, Beutler B, Theofilopoulos AN. Essential requirement for IRF8 and SLC15A4 implicates plasmacytoid dendritic cells in the pathogenesis of lupus. Proc. Natl. Acad. Sci. U. S. A. 2013;110(8):2940–2945. doi: 10.1073/pnas.1222798110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.