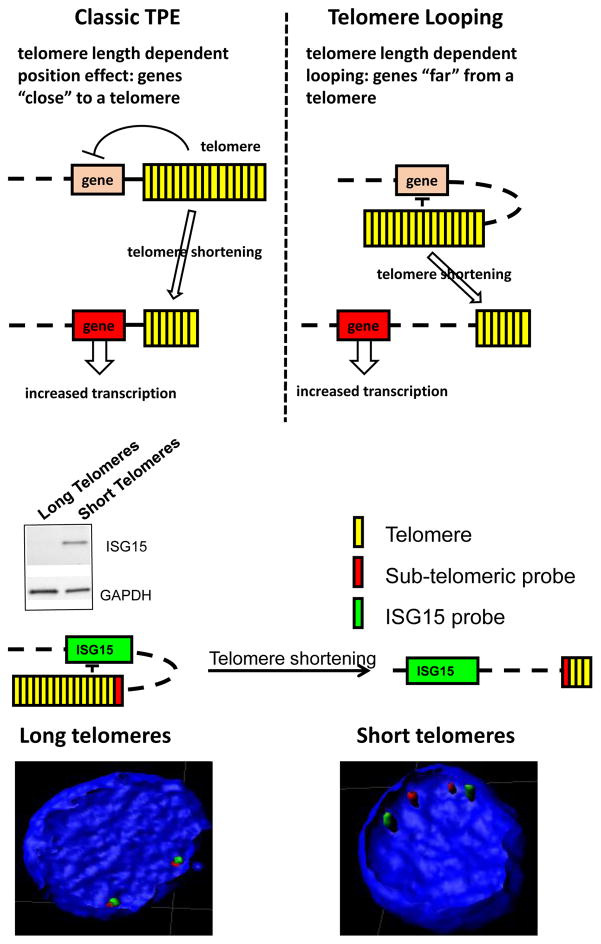
Figure 3.
Comparison of classic telomere position effects (TPE) to telomere looping. TPE (top). In the budding yeast and in mammalian cells TPE is primarily due to deacetylation of subtelomeric nucleosomes. In humans, DUX4, C1S and SORBS2, ISG15, and hTERT expression, some of which are located up to 10 Mb from telomeres can also depend on TPE but by a slightly different mechanism. This novel mechanism involves physical association of the telomere through looping to chromatin (often internal telomere sequences) near these genes and is called TPE-OLD (over long distances). TPE-OLD genes are generally repressed in cells with long telomeres and become expressed in cells with shorter telomeres. Importantly, there are genes between the telomeres and the TPE-OLD regulated genes that are not regulated by telomere length. One example is illustrated for the ISG15 gene (bottom). In young cells with long telomeres ISG15 is repressed but in older cells with short telomere ISG15 is expressed. Increasing telomeres length by introduction of hTERT into old cells with short telomere repressed ISG15 expression but only after substantial telomere re-elongation. In addition to protein expression changes, a subtelomeric probe next to the canonical TTAGGG repeats at the chromosome end are adjacent to the ISG15 locus in young cells with long telomeres while separated in old cells with short telomeres.