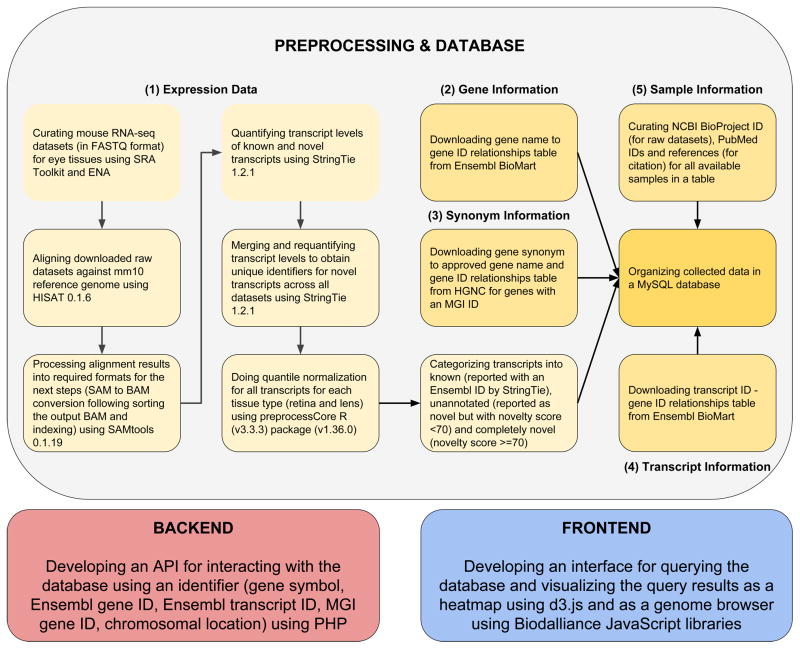
Fig. 1.
Overview of the transcriptome profiling and database construction for Express. Transcriptomes of mouse lens and retina spanning several development stages (with biological replicates) were collected from published sources listed in Tables 1 and 2. Curated RNA sequence data was quality filtered using FASTX Toolkit. High quality raw sequence reads were processed and aligned to mouse reference genome mm10 using HISAT and outputs were collected as Sequence Alignment Map (SAM) files. Post-processing (i.e. conversion of SAM to sorted Binary Alignment Map (BAM)) of aligned reads was accomplished using SAMTools. Aligned and post-processed RNA-seq BAM files associated with each developmental stage were utilized for identifying and quantifying the expression levels of known and novel transcripts across respective development stages of tissue subtypes using StringTie. Quantile normalization was performed for samples per tissue type using preprocess R package. The novel transcripts reported by StringTie were categorized into unannotated (novelty score < 70) and completely novel transcripts (novelty score >= 70). These normalized expression levels of known, unannotated and completely novel transcripts were organized into a table. Gene information mapping gene names to gene IDs was downloaded from Ensembl BioMart. Synonym information mapping gene synonyms to approved gene names and gene IDs was downloaded from Hugo Gene Nomenclature Committee (HGNC) for the genes with an MGI ID. Sample information was manually curated for samples and NCBI BioProject ID, PubMed ID and study reference were obtained per sample. These collected data were then organized into a My Structured Query Language (MySQL) database. Following abbreviations and web resources have been employed in this study: SRA - Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra), ENA - European Nucleotide Archive (https://www.ebi.ac.uk/ena), HISAT - https://ccb.jhu.edu/software/hisat/, SAM - Sequence Alignment Map, BAM - Binary Alignment Map, StringTie - https://ccb.jhu.edu/software/stringtie/, R - https://www.r-project.org/about.html, Ensembl Biomart - https://www.ensembl.org/biomart, HGNC - HUGO Gene Nomenclature Committee, MGI - Mouse Genome Informatics (www.informatics.jax.org), MySQL - My Structured Query Language, API - Application Programming Interface, PHP - Hypertext Preprocessor.