FIGURE 2.
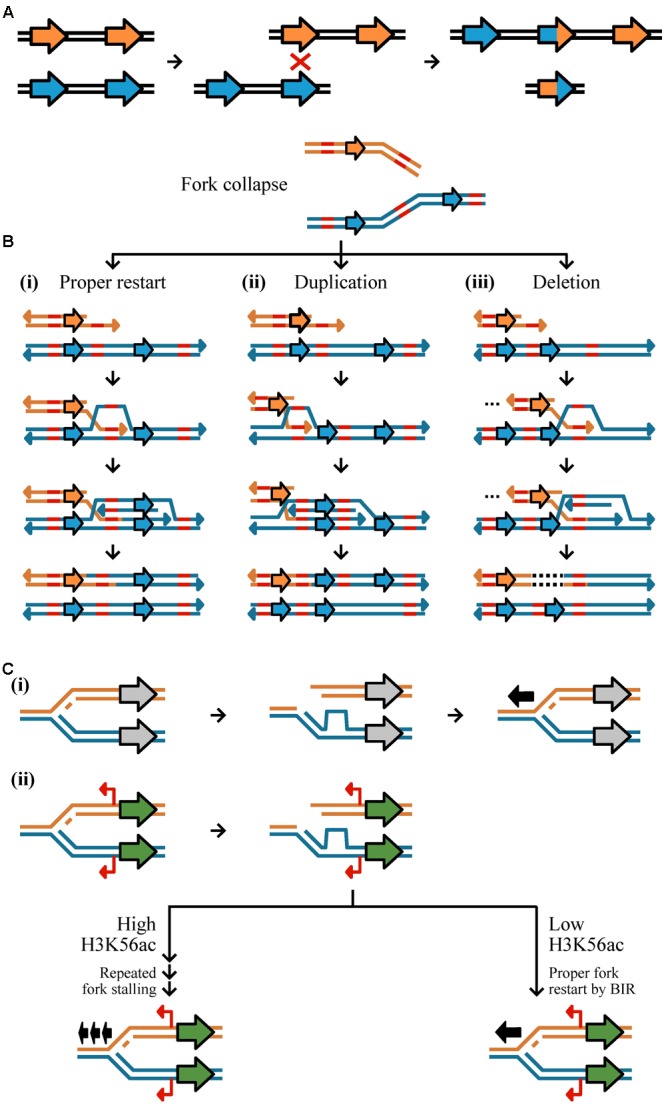
Mechanisms of CN variant formation. CN variants typically occur as a result of aberrant replication via homologous recombination, non-homology based mechanisms, and environmentally stimulated processes. (A) Unequal crossing over during recombination may result in duplication and deletion. Here, two equal strands of DNA with two genes (represented by the orange or blue arrows) have undergone unequal crossing over due to the misalignment of a homologous sequence. This results in one DNA strand having three genes and the other one gene. (B,C) A major driver of CN variant formation is aberrant DNA replication. (B, top) Double strand breaks at replication forks or collapsed forks are often repaired via Break-induced replication (BIR). (i) Proper BIR starts with strand invasion of a homologous or microhomologous sequence (shown in red) to allow for proper fork restart. (ii) If template switching occurs in the backward direction, a segment of DNA will have been replicated twice resulting in a duplication; (iii) in contrast, template switching in the forward direction results in a deletion represented by a dashed line in the DNA sequence. Erroneous BIR may be mediated by microhomologies as well. (C) CN variants may be stimulated near genes that are highly expressed due to an increased chance of fork stalling. (i) If a replication fork breaks down near a gene that is not expressed (gray) and restarts once (represented by one black arrow), no mutation will occur. (ii) If a replication fork breaks down near a gene that is expressed (green) with cryptic unstable transcripts (red) then there may be two outcomes dependent on the degree of the H3K56ac acetylation mark. If there are low levels of H3K56ac, it is more likely that there will be proper fork restart by BIR (represented by one black arrow). If there are high levels of H3K56ac, it is more likely that there will be repeated fork stalling (represented by three black arrows) (see Figure 8 from Hull et al., 2017).
