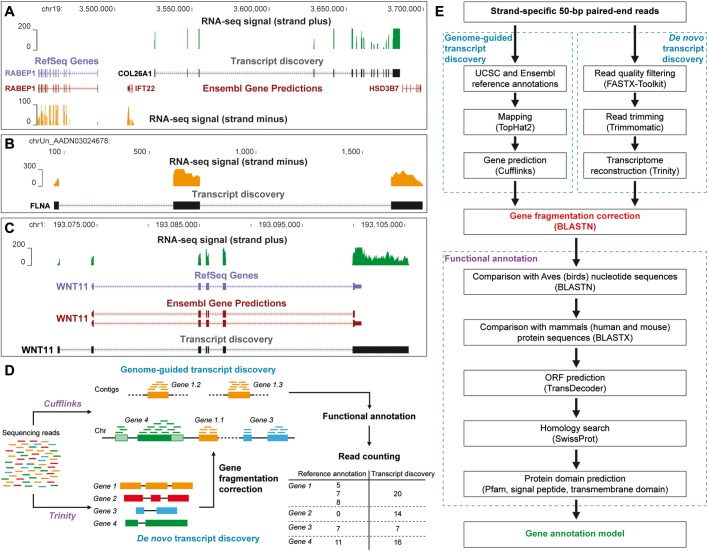
Fig. 1.
Dual transcript-discovery approach. (A) Region surrounding the genes RABEP1 and HSD3B7 on chromosome 19. RNA-seq signal on strand plus (green), which does not overlap any gene from UCSC and Ensembl reference annotations, corresponds to the gene COL26A1. (B) RNA-seq signal (orange) on strand minus of an uncharacterized contig delimitating three exons of the gene FLNA. (C) Region of the gene WNT11 on chromosome 1. As visible from the RNA-seq signal on strand plus (green), both UCSC and Ensembl reference annotations lack an exon of the 5′-UTR and display a shorter 3′-UTR. (D) The dual transcript-discovery approach combined a genome-guided gene prediction with a de novo transcriptome reconstruction. This dual approach enabled us to correct for gene fragmentation (orange), to identify missing gene candidates (red) and to adjust or validate existing annotated genes (green, blue) thus improving the assignment rate of RNA-seq read pairs. (E) Workflow to design the comprehensive gene annotation model.