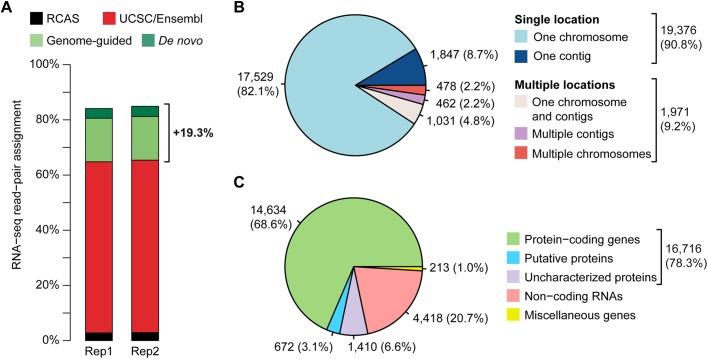
Fig. 2.
Characteristics of the new gene annotation model. (A) The dual transcript-discovery approach combining genome-guided gene prediction (light green) and de novo transcriptome reconstruction (dark green) raised the read-pair assignment rate by 19.3% as compared to when using the UCSC and Ensembl reference annotations (red). The proportion of read pairs coming from the RCAS-BP(A) replication competent retroviruses is depicted in black. (B) Proportion of gene locations on chromosomes and contigs of the chicken reference genome galGal4. Of the identified gene candidates, 9.2% are fragmented due to their location on multiple chromosomes and contigs. (C) Proportion of annotated gene biotypes. Most of the annotated gene candidates potentially encode proteins (78.3%). Putative proteins correspond to gene candidates for which at least one protein domain could be detected (3.1%). Uncharacterized proteins are gene candidates with an ORF of ≥100 amino acids without protein domain identified (6.6%). Gene candidates with no sufficient predicted ORF (<100 amino acids) are classified as non-coding RNAs (20.7%). Gene candidates encoding spliceosome complex members and ribosomal RNAs, as well as pseudogenes, are classified as miscellaneous genes (1.0%).