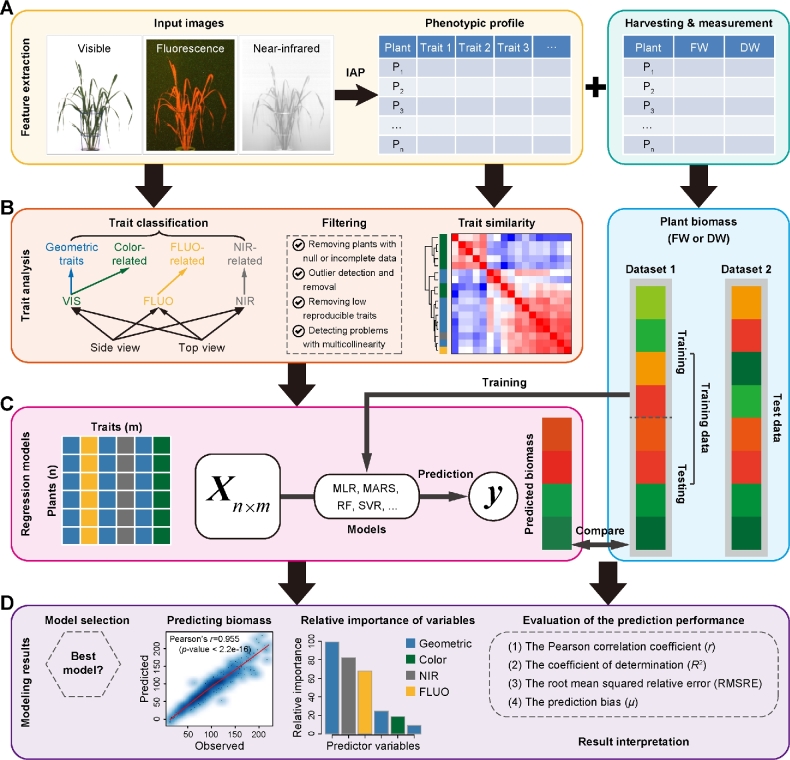
Figure 1:
Modeling pipeline for predicting plant biomass accumulation based on image-derived parameters. A, Input data, including high-throughput image data and manually measured biomass data. Plants were phenotyped using various cameras such as visible (or color), fluorescence, and near-infrared sensors. Image analysis was performed with IAP software [10] for feature extraction. The same plants were harvested and measured at the end of growth. Generally, 2 types of biomass were measured: fresh weight and dry weight. B, Trait processing. All the phenotypic traits were grouped into 4 categories: geometric, color-related, FLUO-related, and NIR-related traits. Phenotypic data were subjected to quality check to remove low-quality data. C, Each plant was described by a list of traits, resulting in a predictor matrix whose rows represent plants and columns represent image-based traits. This matrix was used to predict plant biomass accumulation by MLR, MARS, RF, and SVR models. The right panel represents the schema of model validation. In the first schema, a dataset (Dataset 1) was divided into training set and testing set in a 10-fold cross-validation manner. In the second schema, the whole of 1 dataset (Dataset 1) was used for training and another dataset (Dataset 2) was used for testing. D, Model selection, evaluation, and result interpretation. The correlation of the predicted values and measured values was used to assess the overall performance of the model.