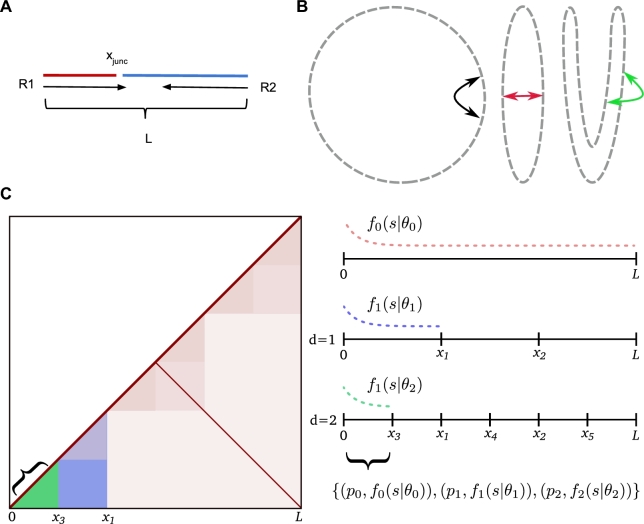
Figure 4:
Model details. Generation of proximity ligation inserts (a) involves joining 2 randomly drawn parts (red and blue), from which the read-pair (R1, R2) is then simulated. The junction point (xjunc) varies over the interval [0..L), and reproduction of read-through events is possible. For an unbounded chromosome (b) (circular here), besides strictly primary separation (black arrow), spatial proximity can be induced from successive folding (red, green arrows). When the spatial arrangement is consistent across the population of cells, this will be observable as modulations in the contact frequencies. Sim3C models simple structurally related modulation of observed contact frequencies (c). Beyond primary interactions forming the main diagonal, users can reproduce inter-arm-mediated anti-diagonals. Finer-scale modulations attributed to topologically associated domains can optionally be randomly simulated. Primary interactions f0(s|θ0) (equation 1) cover the full interval [0, L). Each level of recursion (d = 1, 2…n) generates a finer set of intervals, to which a distribution fi(s|θi) and probability pi are assigned. The final covering of intervals each define a range (green, curly braces) over which a set of probabilities and empirical distribution pairs govern interaction separation s.