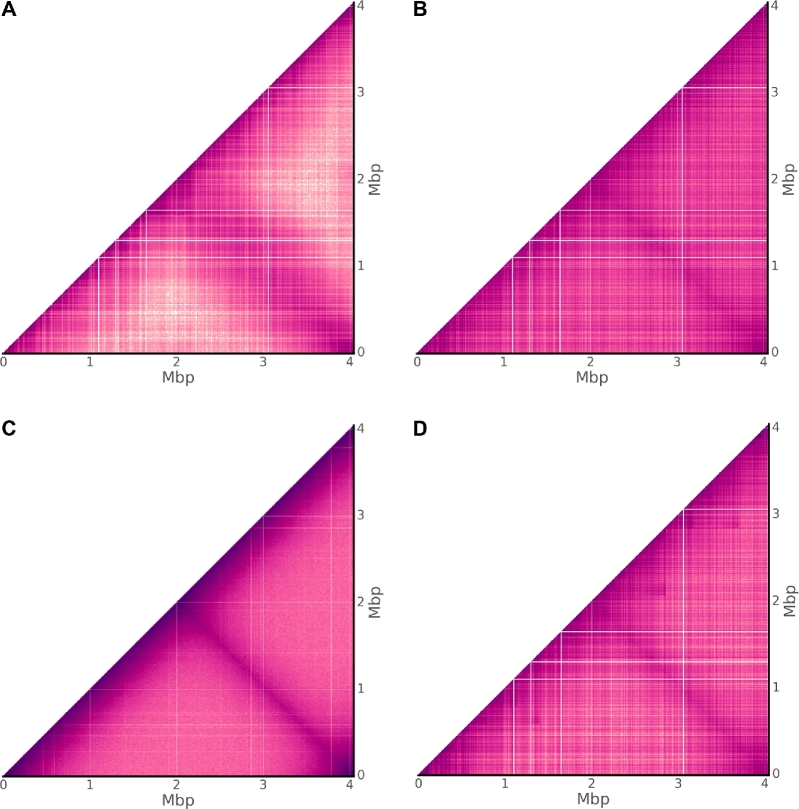
Figure 5:
Bacterial contact maps. Observed Hi-C interactions for the monochromosomal genome of Caulobacter crescentus NA1000. Comparing (a) real experimental data [26] with the 3 simulation choices (b) traditional Hi-C, (c) DNase Hi-C, and (d) traditional Hi-C with TADs enabled. Sharp rectilinear modulations of the intensity within (a) and (b) indicate a reduction in PL observations within a given bin. Not due to 3D chromosome structure, rather such features can be attributed largely to mappability and low cut-site density. (c) Without an enzymatic constraint, a significantly smoother field is apparent, yet still susceptible to mappability. (d) Enabling topologically associated domains highlights the similarity between features produced merely from biases and what could be truly associated with 3D structure.