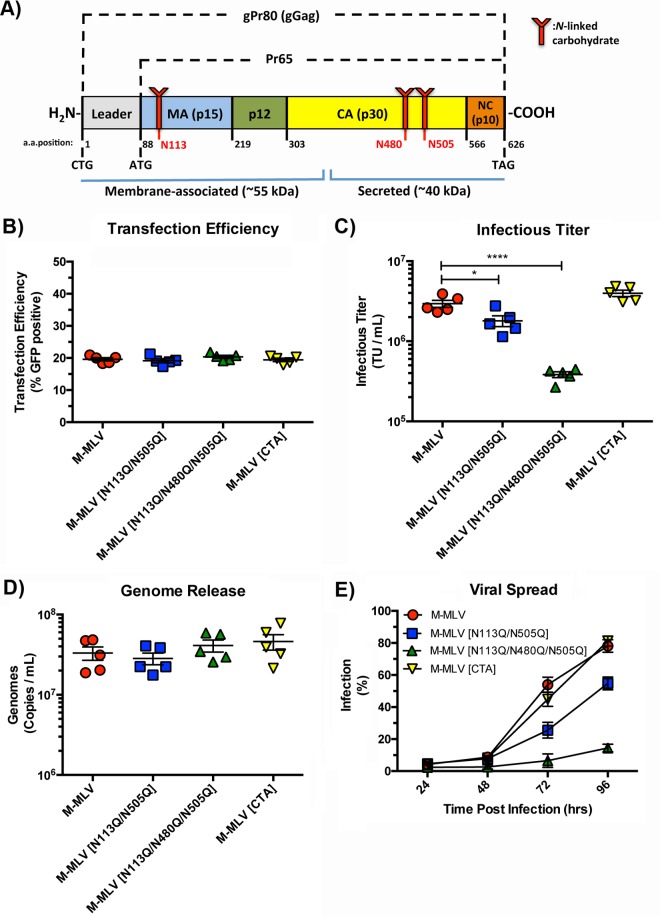
FIG 1.
Replicative fitness of gPr80-deficient and mutant viruses. (A) Schematic representation of the gPr80 and Pr65 proteins. The locations of the three N-linked glycosylation sites are indicated in red. The approximate regions encompassing the N-terminal membrane-associated domain and the C-terminal secreted domain are indicated with blue lines; the leader region contains the ER localization signal peptide. (B) Transfection efficiencies of virus expression constructs in transfected 293T cells. (C) Infectious titers of viruses released from transfected 293T cells. Viruses were harvested after 48 h and filtered through 0.45-μm filters. Equal volumes of the different viruses were then titrated on NIH 3T3 cells, and infection was measured by flow cytometry 24 h after infection. (D) Viral genomes released into the supernatants of the same transfected 293T cells as those described above were measured by digital droplet PCR (ddPCR). Samples were treated with DNase I prior to RNA extraction. (E) Replicative fitness of M-MLV and its glycosylation mutants. Input virus was normalized via the number of TU per milliliter. Infection was monitored by measuring EGFP expression at each time point following infection by flow cytometry. Data are the compilation of five independent transfection experiments done in triplicate. P values were calculated by Student's t test. *, P ≤ 0.05; ****, P ≤ 0.0001. P values for the data sets in panels B and D were not significant for comparisons to the M-MLV control (ns, P > 0.05).