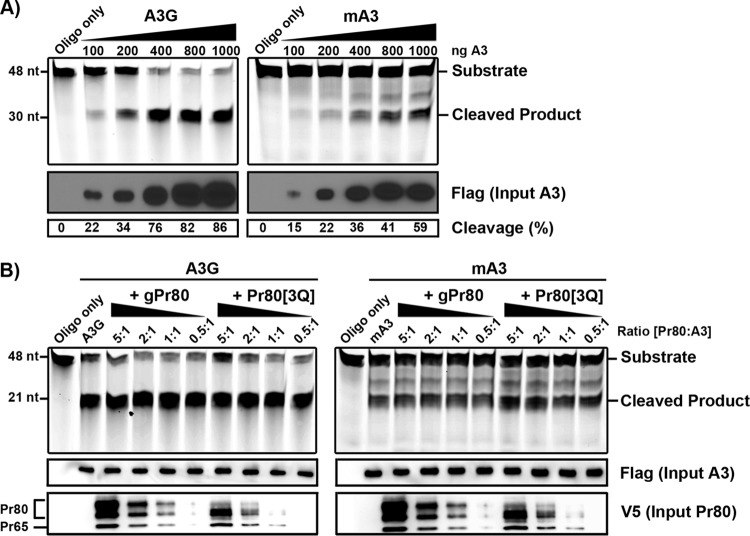
FIG 6.
Oligonucleotide deamination assays. (A) Oligonucleotide deamination was performed using purified mA3 and A3G. Lysates of 293T cells expressing FLAG-tagged mA3 or A3G were precipitated with anti-FLAG-conjugated agarose beads. Proteins were eluted from the beads and then used in oligonucleotide cleavage assays with 0.1 pmol/μl of FAM-labeled oligonucleotides containing TTC or CCC target sites. Deaminated oligonucleotides were treated with uracil DNA glycosylase (UDG), boiled, and resolved in an acrylamide gel. Input A3 protein and the efficiency of oligonucleotide cleavage are shown below the input gels. The percentage of cleaved oligonucleotide in each sample was determined using ImageJ software. (B) Oligonucleotide deamination assays in the presence of purified gPr80-V5 or Pr80[3Q]-V5 and 800 ng of mA3 or A3G were carried out as described for panel A. Molar ratios of gPr80/Pr80[3Q] to A3 are indicated above the gels. Input gPr80/Pr80[3Q] was assessed using a polyclonal anti-p30 antibody. The average cleavage efficiency was 84% for A3G and 46% for mA3 under all conditions in two independent experiments.