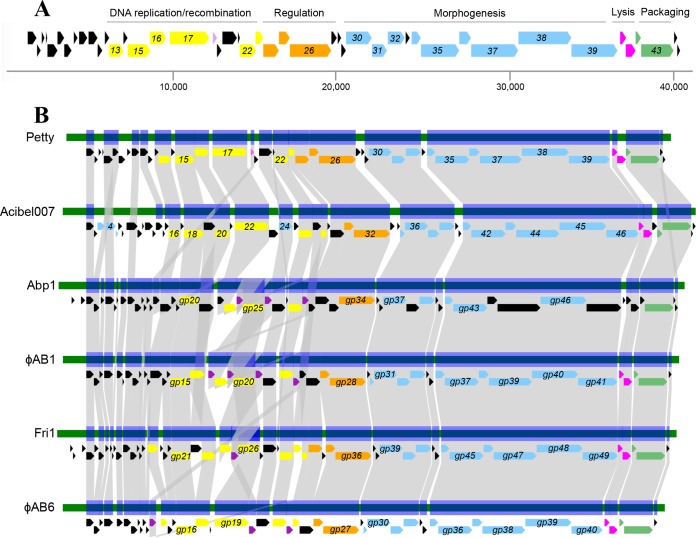
FIG 3.
Genome architecture of phage Petty and other ϕKMV-like Acinetobacter phages. (A) Genome architecture and predicted ORFs of phage Petty are shown. The linear map is based on nucleotide sequences of the phage genome and predicted ORFs. ORFs are color-coded based on their functional role category: yellow for phage DNA replication and recombination, orange for regulation, purple for endonucleases, blue for morphogenesis-related proteins, pink for lysis, and green for DNA packaging. Unattributed genes are colored black. Selected gene numbers are shown. Like for other ϕKMV-like phages, the Petty RNA polymerase gene is adjacent to the morphogenesis genes. Interestingly, the lysis cassette of the phage genomes does not have a predicted canonical spanin. (B) Genome maps and protein identity comparison between selected ϕKMV-like phages infecting Acinetobacter. Phages are aligned relative to gp1, which is highly conserved in this genus. Gene products (gp) sharing significant sequence identity are linked in gray (see Materials and Methods).