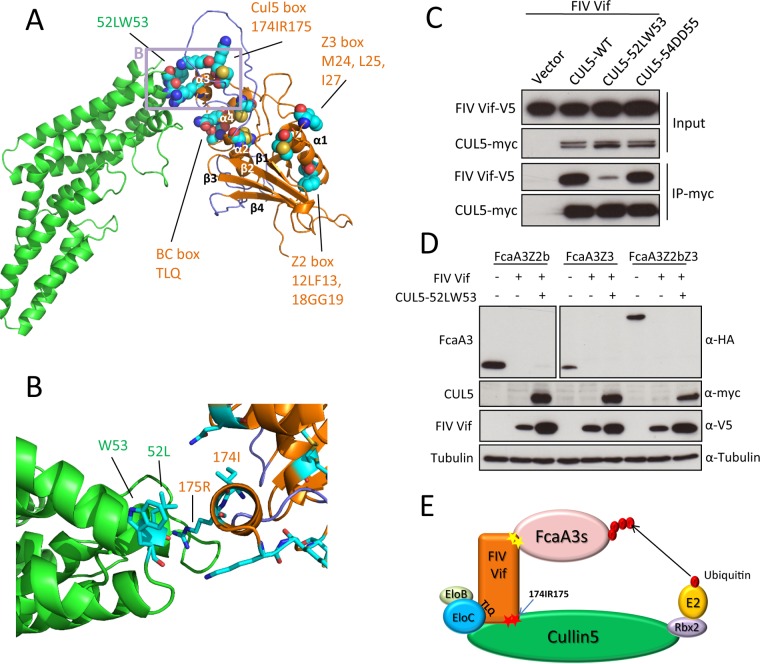
FIG 5.
FIV Vif-CUL5 three-dimensional structure model. (A) Homology model of the FIV Vif (orange)-CUL5 (green) complex in a cartoon representation. Helices α3 and α4 of Vif are interacting with CUL5. The model contains two unstructured loops (navy) before and after helix α2, as no structural template is available for these regions. These loops might be important for binding other parts of the complex. Residues that were subjected to mutational analysis are shown in a sphere representation. The region of the closeup shown in panel B is indicated by a light plum rectangle. (B) Closeup view of the homology model of the FIV Vif (orange)-CUL5 (green) complex in a cartoon representation, with interacting residues shown in a stick representation. (C) Expression plasmids for wild-type myc-CUL5 or the indicated mutants were cotransfected with the expression plasmid for wild-type FIV Vif-V5 into HEK293T cells. Cell lysates were immunoprecipitated with anti-myc beads and then analyzed by immunoblotting with anti-V5 antibody for FIV Vif and anti-myc antibody for CUL5. (D) HEK293T cells were cotransfected with expression plasmids for FcaA3Z2b-HA, FcaA3Z3-HA, or FcaA3Z2bZ3-HA and the CUL5 52LW53-AA mutant with FIV Vif-V5. pcDNA3.1 was used as a control plasmid to replace the FIV Vif or CUL5 expression plasmids. Cells were analyzed by immunoblotting using anti-HA, anti-V5, anti-CUL5, anti-Flag, and antitubulin antibodies. (E) Model of FIV Vif with the E3 ligase complex. The CUL5 interaction sites of FIV Vif (174IR175) are shown as red stars.