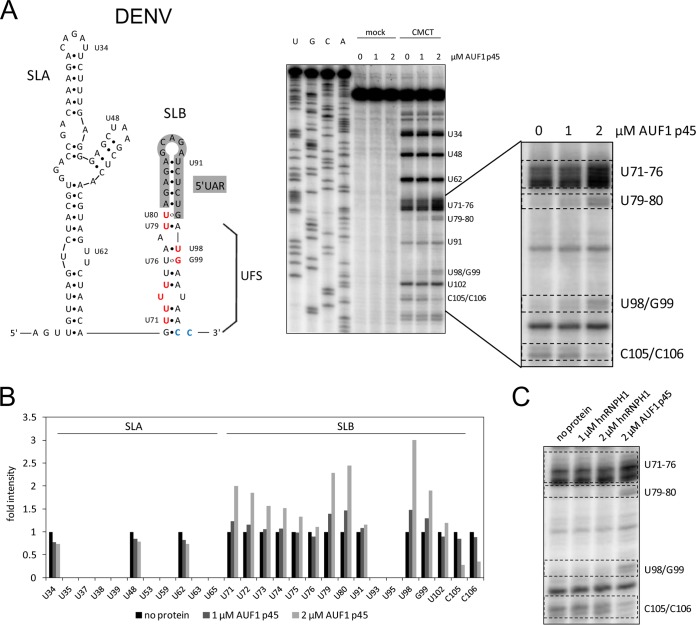
FIG 4.
AUF1 p45 specifically destabilizes SLB within the DENV 5′UTR. (A) (Left) Secondary structures of 5′-terminal stem-loop A (SLA) and stem-loop B (SLB) of the DENV genome. Nucleotide position numbering starts at the 5′ end of the RNA. The 5′UAR-flanking stem (UFS) is indicated. Nucleotides that are exposed by AUF1 p45 are highlighted in red. Nucleotides that are less well modified by CMCT (modifies U and G residues in single-stranded RNA) in the presence of AUF1 p45 are highlighted in blue. (Right) RNA structure probing of the DENV 5′ end by CMCT in the presence of the indicated concentrations of AUF1 p45 (lanes 8 to 10). Control reaction mixtures in CMCT buffer were included (lanes 5 to 7). Products of the primer extension reaction, carried out with a radiolabeled primer, were analyzed by 8% denaturing PAGE along with a sequencing ladder (lanes 1 to 4). Note that due to the stop of the reverse transcriptase upon encountering alkylated Watson-Crick positions, the resulting product is one base shorter. One region of the gel is enlarged on the right. (B) Quantification of the primer extension analysis shown in panel A. (C) RNA structure probing of the DENV 5′ end with CMCT in the presence of the indicated concentrations of AUF1 p45 and hnRNPH1. One region of the gel where we observed a change of the secondary structure in the presence of AUF1 p45 is shown (see panel A).